Submitted:
06 November 2024
Posted:
07 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
- (1)
- the differential rate of newly infected persons from the deseasewhich, with a delay time of about a week, determines the death rate , where the mortality rate f is of the order – varying for different mutants of the Covid virus and different countries [102]. also determines the hospitalization rate of seriously infected persons.
- (2)
- the fraction of infected persons determines the peak time of required clinical resources in the host country of the considered population [103].
2. Reduction of the General SIR-Equations
2.1. Reduction
2.2. Stationary Ratio
3. Exact Solution
3.1. Ansatz
3.2. Resulting Fractions
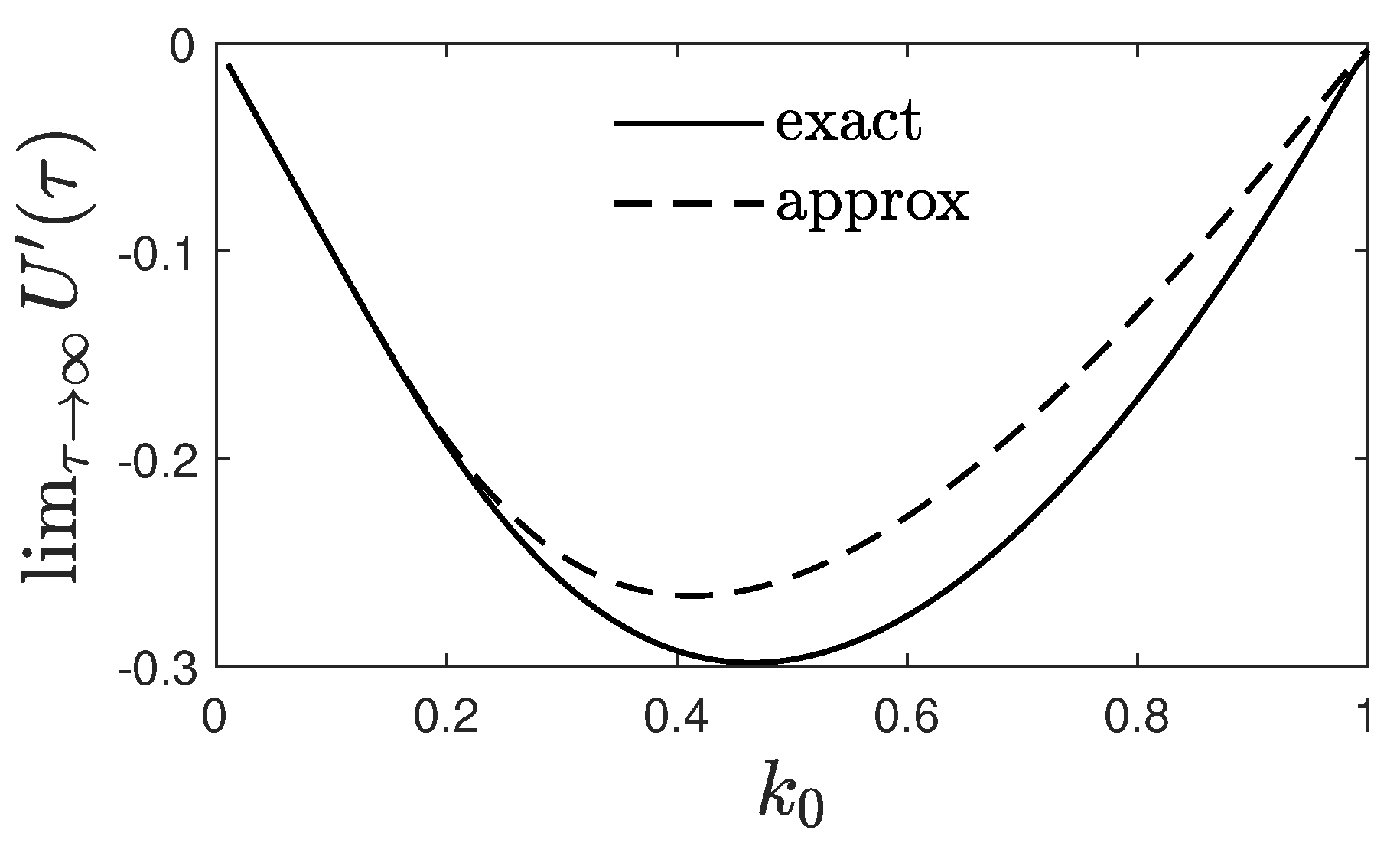
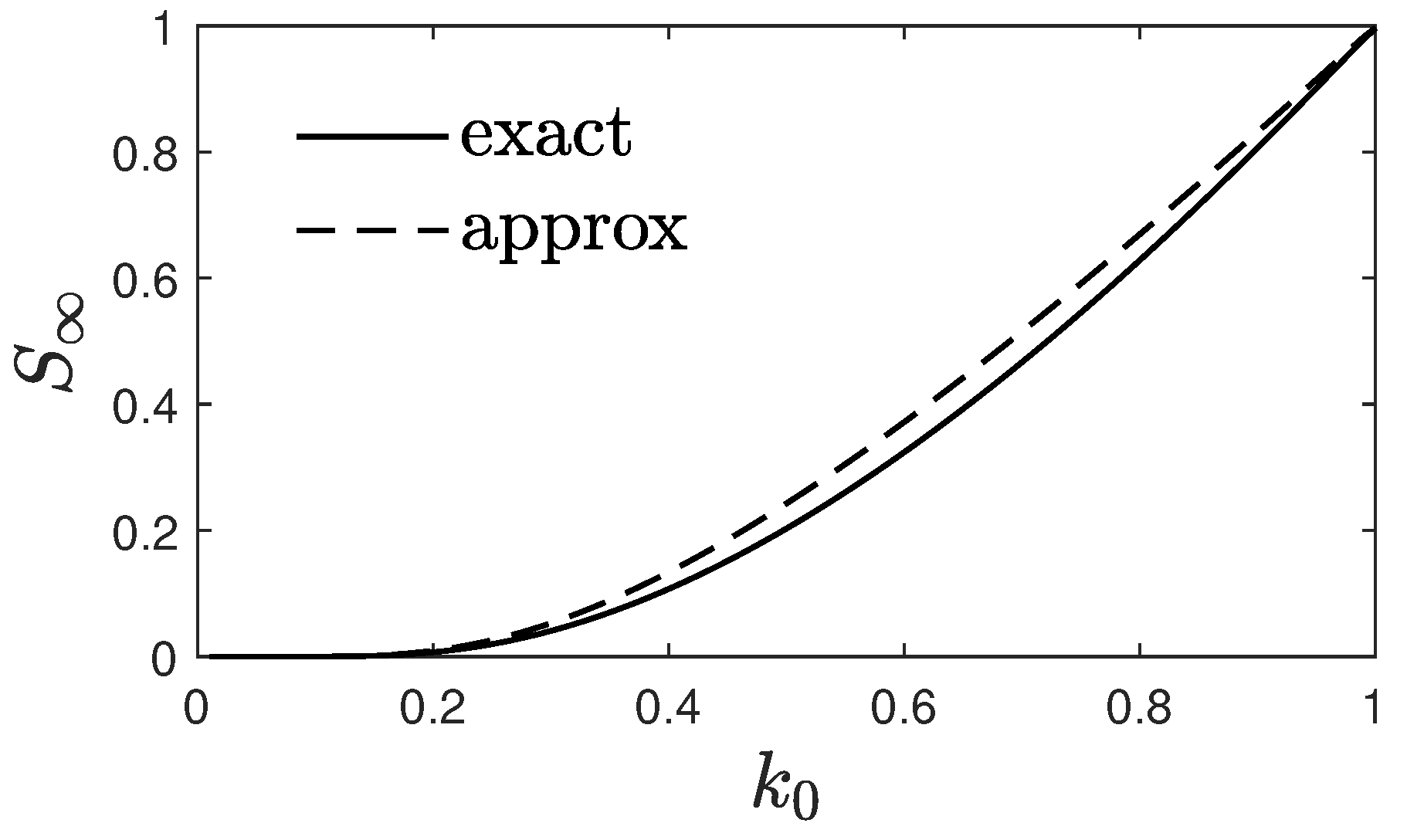
4. Approximated for Large and Small Times
4.1. Large Times
4.2. Small Times
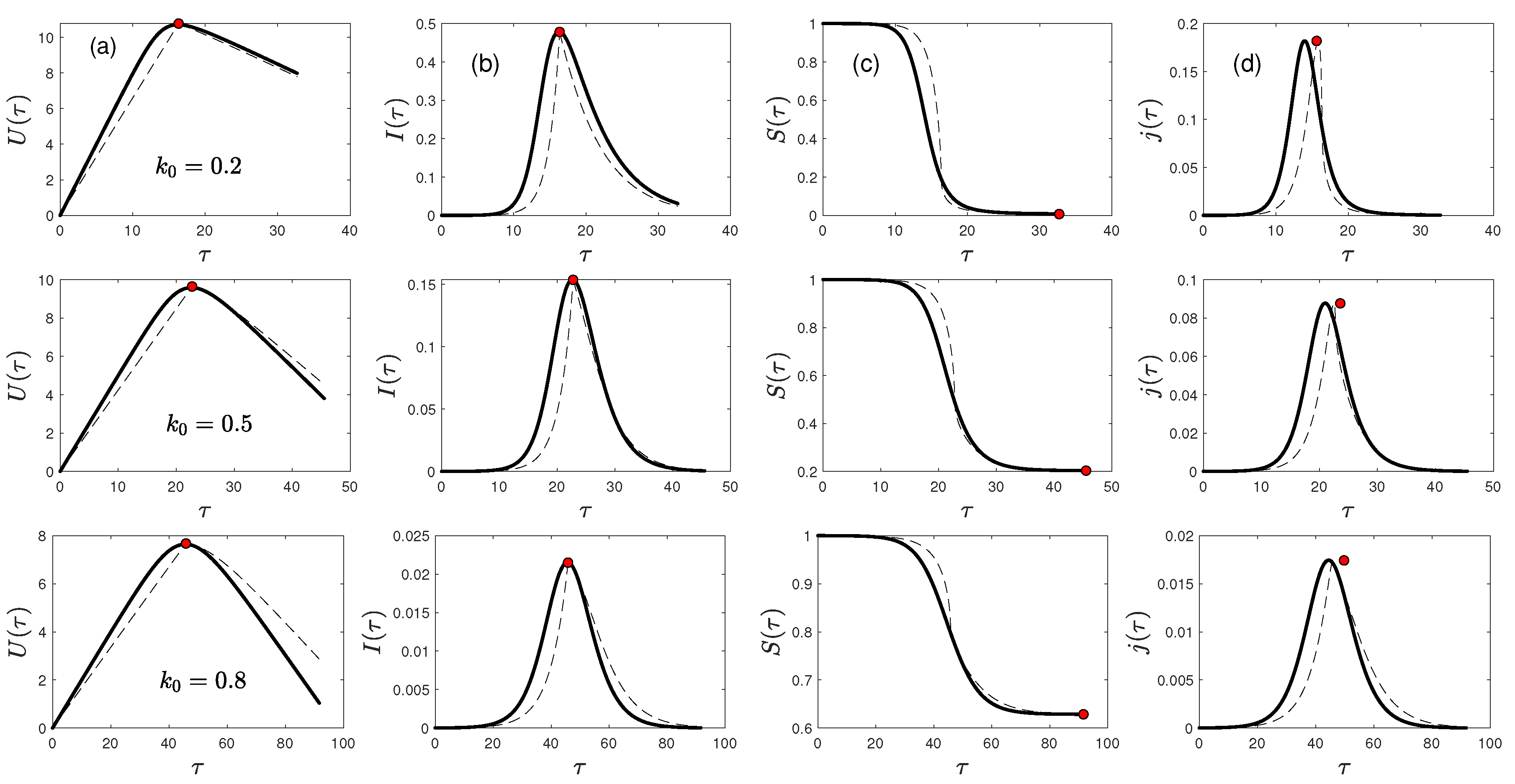
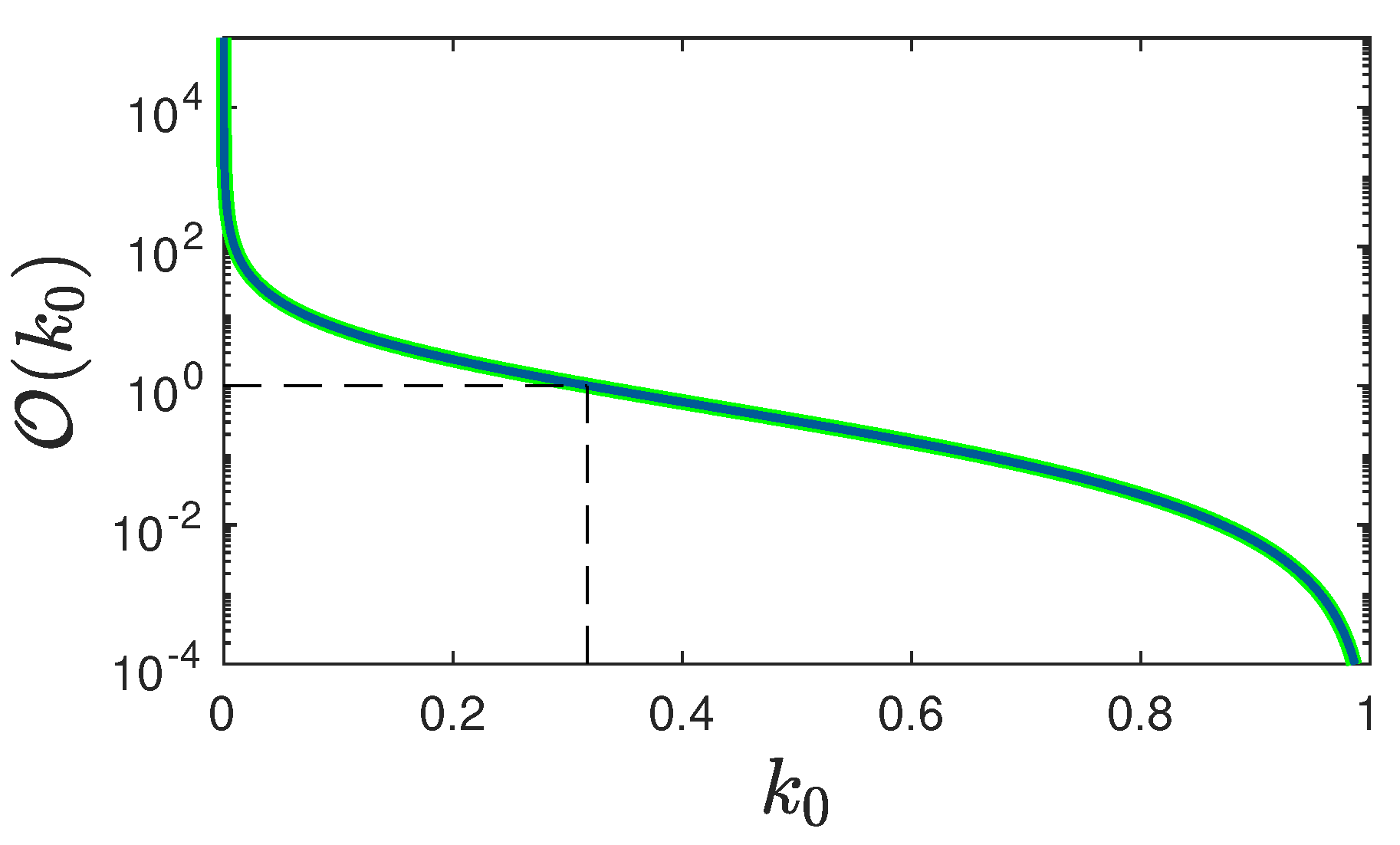
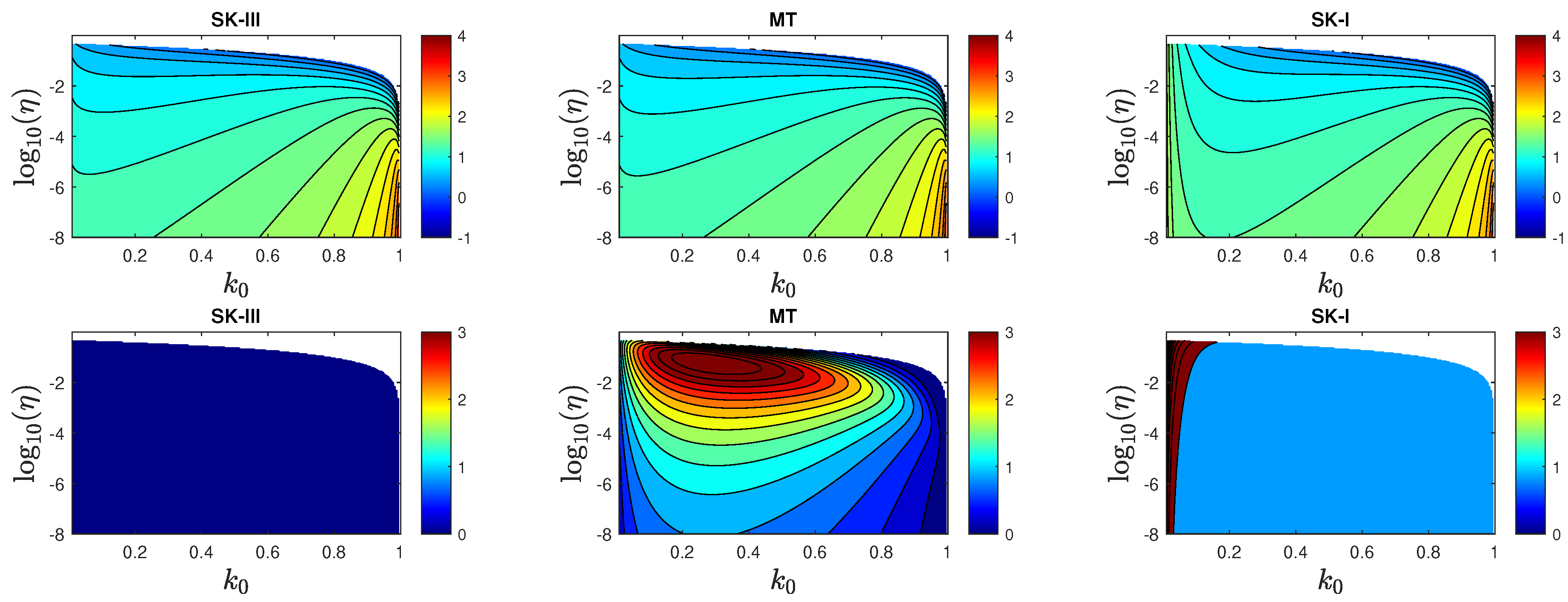
5. Results
5.1. Rate of New Infections
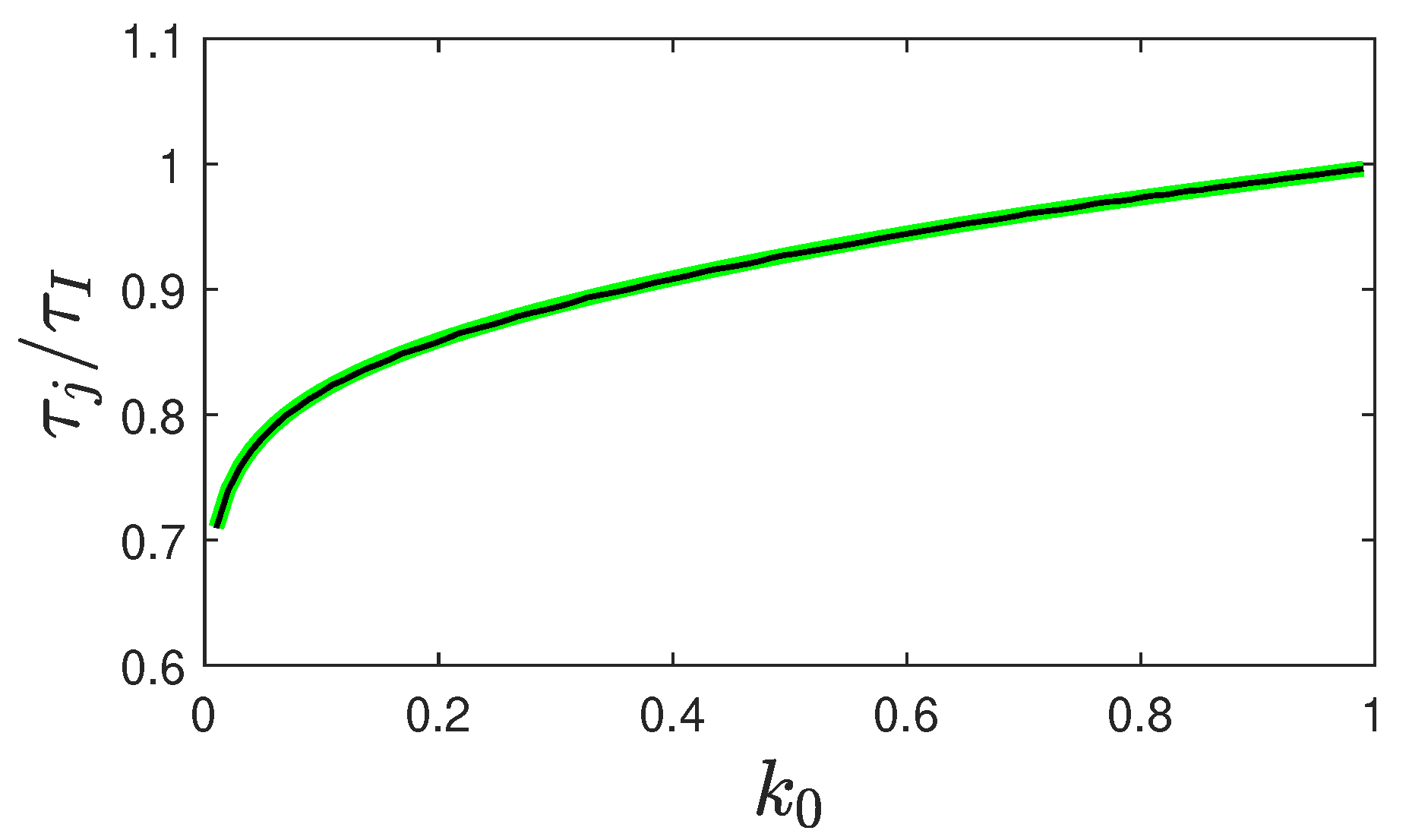
5.2. Peak Time of Fraction of Infected Persons
6. Summary and Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
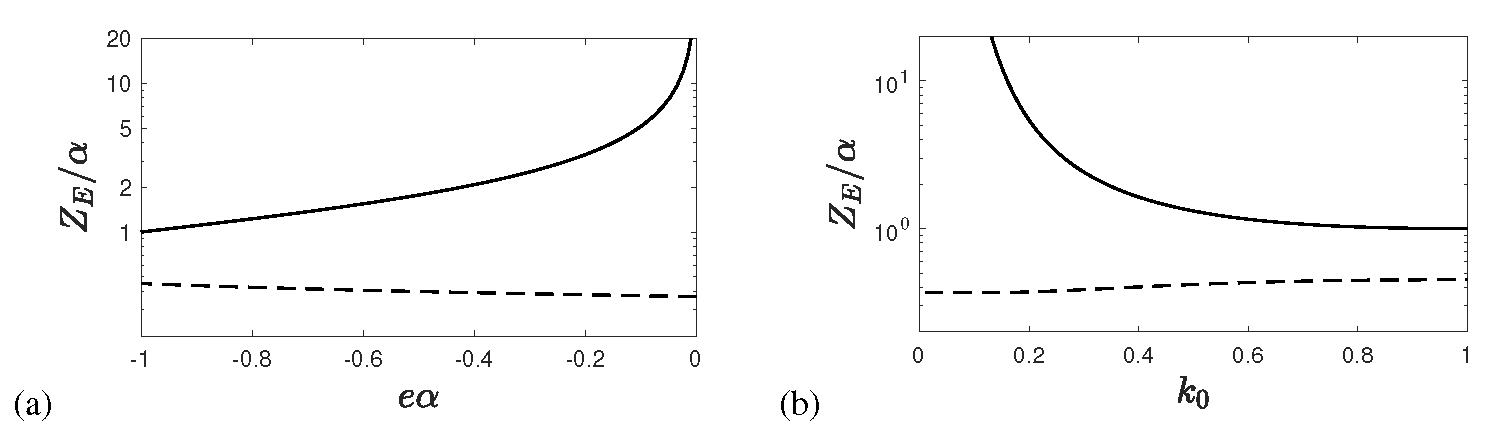
Appendix A. Inverse KSSIR-Solution of Earlier Work
Appendix A.1. Final and Maximum Values
Appendix A.2. Peak Differential Rate
References
- Kermack, W.O.; McKendrick, A.G. A contribution to the mathematical theory of epidemics. Proc. Royal Soc. A 1927, 115, 700–721. [Google Scholar]
- Kendall, D.G. Deterministic and stochastic epidemics in closed populations. In Proc. Third Berkeley Symp. on Math. Statist. and Prob.; Univ. of Calif. Press, Berkeley, United States, 1956; Vol. 4, pp. 149–165.
- Babajanyan, S.G.; Cheong, K.H. Age-structured SIR model and resource growth dynamics: a Covid-19 study. Nonlinear Dynamics 2021, 104, 2853–2864. [Google Scholar] [CrossRef] [PubMed]
- Turkyilmazoglu, M. Explicit formulae for the peak time of an epidemic from the SIR model. Physica D-nonlinear Phenomena 2021, 422, 132902. [Google Scholar] [CrossRef] [PubMed]
- Telles, C.R.; Lopes, H.; Franco, D. SARS-COV-2: SIR Model Limitations and Predictive Constraints. Symmetry-basel 2021, 13, 676. [Google Scholar] [CrossRef]
- Carvalho, A.M.; Goncalves, S. An analytical solution for the Kermack-McKendrick model. Physica A 2021, 566, 125659. [Google Scholar] [CrossRef]
- Priesemann, V.; Brinkmann, M.M.; Ciesek, S.; Cuschieri, S.; Czypionka, T.; Giordano, G.; Gurdasani, D.; Hanson, C.; Hens, N.; Iftekhar, E.; Kelly-Irving, M.; Klimek, P.; Kretzschmar, M.; Peichl, A.; Perc, M.; Sannino, F.; Schernhammer, E.; Schmidt, A.; Staines, A.; Szczurek, E. Calling for pan-European commitment for rapid and sustained reduction in Sars-Cov-2 infections. Lancet 2021, 397, 92–93. [Google Scholar] [CrossRef]
- Mungkasi, S. Variational iteration and successive approximation methods for a SIR epidemic model with constant vaccination strategy. Applied Mathematical Modelling 2021, 90, 1–10. [Google Scholar] [CrossRef]
- dos Santos, I.F.F.; Almeida, G.M.A.; de Moura, F.A.B.F. Adaptive SIR model for propagation of Sars-Cov-2 in Brazil. Physica A 2021, 569, 125773. [Google Scholar] [CrossRef]
- Ghaffar, A.; Alanazi, S.; Alruwaili, M.; Sattar, M.U.; Ali, W.; Humayun, M.; Siddiqui, S.Y.; Ahmad, F.; Khan, M.A. Multi-Stage Intelligent Smart Lockdown using SIR Model to Control Covid 19. Intelligent Automation And Soft Computing 2021, 28, 429–445. [Google Scholar] [CrossRef]
- Law, K.B.; Peariasamy, K.M.; Gill, B.S.; Singh, S.; Sundram, B.M.; Rajendran, K.; Dass, S.C.; Lee, Y.L.; Goh, P.P.; Ibrahim, H.; Abdullah, N.H. Tracking the early depleting transmission dynamics of Covid-19 with a time-varying SIR model. Scientific Reports 2020, 10, 21721. [Google Scholar] [CrossRef]
- Brugnano, L.; Iavernaro, F.; Zanzottera, P. A multiregional extension of the SIR model, with application to the Covid-19 spread in Italy. Mathematical Methods In The Applied Sciences 2021, 44, 4414–4427. [Google Scholar] [CrossRef]
- Venkatasen, M.; Mathivanan, S.K.; Jayagopal, P.; Mani, P.; Rajendran, S.; Subramaniam, U.; Ramalingam, A.C.; Rajasekaran, V.A.; Indirajithu, A.; Sorakaya Somanathan, M. Forecasting of the Sars-Cov-2 epidemic in India using SIR model, flatten curve and herd immunity. Journal of Ambient Intelligence And Humanized Computing 2020. [Google Scholar] [CrossRef] [PubMed]
- Heng, K.; Althaus, C.L. The approximately universal shapes of epidemic curves in the Susceptible-Exposed-Infectious-Recovered (SEIR) model. Scientific Reports 2020, 10, 19365. [Google Scholar] [CrossRef] [PubMed]
- Chekroun, A.; Kuniya, T. Global threshold dynamics of aninfection age-structured SIR epidemic model with diffusion under the Dirichlet boundary condition. Journal of Differential Equations 2020, 269, 117–148. [Google Scholar] [CrossRef]
- Cadoni, M.; Gaeta, G. Size and timescale of epidemics in the SIR framework. Physica D-nonlinear Phenomena 2020, 411, 132626. [Google Scholar] [CrossRef]
- Chen, Y.C.; Lu, P.E.; Chang, C.S.; Liu, T.H. A Time-Dependent SIR Model for Covid-19 With Undetectable Infected Persons. Ieee Transactions On Network Science And Engineering 2020, 7, 3279–3294. [Google Scholar] [CrossRef]
- Cooper, I.; Mondal, A.; Antonopoulos, C.G. A SIR model assumption for the spread of Covid-19 in different communities. Chaos Solitons Fract. 2020, 139, 110057. [Google Scholar] [CrossRef]
- Karaji, P.T.; Nyamoradi, N. Analysis of a fractional SIR model with General incidence function. Appl. Math. Lett. 2020, 108, 106499. [Google Scholar] [CrossRef]
- McMahon, A.; Robb, N.C. Reinfection with Sars-Cov-2: Discrete SIR (Susceptible, Infected, Recovered) Modeling Using Empirical Infection Data. Jmir Public Health And Surveillance 2020, 6, 279–287. [Google Scholar] [CrossRef]
- Chen, X.; Li, J.; Xiao, C.; Yang, P. Numerical solution and parameter estimation for uncertain SIR model with application to Covid-19. Fuzzy Optimization And Decision Making 2021, 20, 189–208. [Google Scholar] [CrossRef]
- Ahmetolan, S.; Bilge, A.H.; Demirci, A.; Peker-Dobie, A.; Ergonul, O. What Can We Estimate From Fatality and Infectious Case Data Using the Susceptible-Infected-Removed (SIR) Model? A Case Study of Covid-19. Front. Med. 2020, 7, 556366. [Google Scholar] [CrossRef] [PubMed]
- Gopagoni, D.; Lakshmi, P., V. Susceptible, Infectious and Recovered (SIR Model) Predictive Model to Understand the Key Factors of Covid-19 Transmission. International Journal of Advanced Computer Science And Applications 2020, 11, 296–302. [Google Scholar] [CrossRef]
- Sene, N. SIR epidemic model with Mittag?Leffler fractional derivative. Chaos Solitons Fract. 2020, 137, 109833. [Google Scholar] [CrossRef]
- Mohamadou, Y.; Halidou, A.; Kapen, P.T. A review of mathematical modeling, artificial intelligence and datasets used in the study, prediction and management of Covid-19. Applied Intelligence 2020, 50, 3913–3925. [Google Scholar] [CrossRef]
- Barlow, N.S.; Weinstein, S.J. Accurate closed-form solution of the SIR epidemic model. Physica D-nonlinear Phenomena 2020, 408, 132540. [Google Scholar] [CrossRef]
- Simon, M. SIR epidemics with stochastic infectious periods. Stochastic Processes And Their Applications 2020, 130, 4252–4274. [Google Scholar] [CrossRef]
- Postnikov, E.B. Estimation of Covid-19 dynamics "on a back-of-envelope?: Does the simplest SIR model provide quantitative parameters and predictions? Chaos Solitons Fract. 2020, 135, 109841. [Google Scholar] [CrossRef]
- Shah, S.T.A.; Mansoor, M.; Mirza, A.F.; Dilshad, M.; Khan, M.I.; Farwa, R.; Khan, M.A.; Bilal, M.; Iqbal, H.M.N. Predicting Covid-19 Spread in Pakistan using the SIR Model. Journal of Pure And Applied Microbiology 2020, 14, 1423–1430. [Google Scholar] [CrossRef]
- Zhao, X.; He, X.; Feng, T.; Qiu, Z. A stochastic switched SIRS epidemic model with nonlinear incidence and vaccination: Stationary distribution and extinction. International Journal of Biomathematics 2020, 13, 2050020. [Google Scholar] [CrossRef]
- Richard, N.A.; Robert, D.; Valentin, D.; Emma, H.B.; Jan, A. Potential impact of seasonal forcing on a Sars-Cov-2 pandemic. Swiss Medical Weekly 2020, 150, w20224. [Google Scholar]
- Colombo, R.M.; Garavello, M. Optimizing vaccination strategies in an age structured SIR model. Mathematical Biosciences And Engineering 2020, 17, 1074–1089. [Google Scholar] [CrossRef] [PubMed]
- Samanta, S.; Sahoo, B.; Das, B. Dynamics of an epidemic system with prey herd behavior and alternative resource to predator. Journal of Physics A 2019, 52, 425601. [Google Scholar] [CrossRef]
- El Koufi, A.; Adnani, J.; Bennar, A.; Yousfi. Analysis of a Stochastic SIR Model with Vaccination and Nonlinear Incidence Rate. International Journal of Differential Equations 2019, 2019, 9275051. [Google Scholar] [CrossRef]
- Li, X.; Li, X.; Zhang, Q. Time to extinction and stationary distribution of a stochastic susceptible-infected-recovered-susceptible model with vaccination under. Mathematical Population Studies 2020, 27, 259–274. [Google Scholar] [CrossRef]
- Imron, C.; Hariyanto. ; Yunus, M.; Surjanto, S.D.; Dewi, N.A.C. Stability and persistence analysis on the epidemic model multi-region multi-patches. International Conference On Mathematics: Pure, Applied And Computation 2019, 1218, 012035. [Google Scholar]
- Britton, T.; Pardoux, E., Eds. Stochastic Epidemic Models with Inference; Vol. 2255, Springer, Berlin, 2019; pp. 1–472.
- Zhang, Y.; Li, Y.; Zhang, Q.; Li, A. Behavior of a stochastic SIR epidemic model with saturated incidence and vaccination rules. Physica A 2018, 501, 178–187. [Google Scholar] [CrossRef]
- Xu, C.; Li, X. The threshold of a stochastic delayed SIRS epidemic model with temporary immunity and vaccination. Chaos Solitons Fract. 2018, 111, 227–234. [Google Scholar] [CrossRef]
- Liu, Q.; Jiang, D.; Shi, N.; Hayat, T. Dynamics of a stochastic delayed SIR epidemic model with vaccination and double diseases driven by Levy jumps. Physica A 2018, 492, 2010–2018. [Google Scholar] [CrossRef]
- Witbooi, P.J. Stability of a Stochastic Model of an SIR Epidemic with Vaccination. Acta Biotheoretica 2017, 65, 151–165. [Google Scholar] [CrossRef]
- Miller, J.C. Mathematical models of SIR disease spread with combined non-sexual and sexual transmission routes. Infectious Disease Modelling 2017, 2, 35–55. [Google Scholar] [CrossRef]
- Jornet-Sanz, M.; Corberan-Vallet, A.; Santonja, F.J.; Villanueva, R.J. A Bayesian stochastic SIRS model with a vaccination strategy for the analysis of respiratory syncytial virus. Sort-statistics And Operations Research Transactions 2017, 41, 159–175. [Google Scholar]
- Estrada, E. Covid-19 and Sars-Cov-2, Modeling the present, looking at the future. Phys. Rep. 2020, 869, 1. [Google Scholar] [CrossRef] [PubMed]
- Schlickeiser, R.; Kröger, M. Determination of a key pandemic parameter of the SIR-epidemic model from past Covid-19 mutant waves and its variation for the validity of the Gaussian evolution. Physics 2023, 5, 205–214. [Google Scholar] [CrossRef]
- Schlickeiser, R.; Kröger, M. Analytical solution of the Susceptible-Infected-Recovered/Removed model for the not too late temporal evolution of epidemics for general time-dependent recovery and infection rates. Covid 2023, 3, 1781–1796. [Google Scholar] [CrossRef]
- Kröger, M.; Schlickeiser, R. Verification of the accuracy of the SIR model in forecasting based on the improved SIR model with a constant ratio of recovery to infection rate by comparing with monitored second wave data. R. Soc. Open Sci. 2021, 8, 211379. [Google Scholar] [CrossRef]
- Schlickeiser, R.; Kröger, M. Analytical solution of the SIR-model for the temporal evolution of epidemics. Part B. Semi-time case. J. Phys. A: Math. Theor. 2021, 54, 175601. [Google Scholar] [CrossRef]
- Guo, X.; Wang, J.L. A Unified Adsorption Kinetic Model Inspired by Epidemiological Model: Based on Adsorbates "Infect" Adsorbents. Langmuir 2024, 40, 15569–15579. [Google Scholar] [CrossRef]
- Guo, X.; Wang, J.L. A novel monolayer adsorption kinetic model based on adsorbates "infect" adsorbents inspired by epidemiological model. Water Research 2024, 253, 121313. [Google Scholar] [CrossRef]
- Cui, S.X.; Liu, F.Z.; Jardon-Kojakhmetov, H.; Cao, M. Discrete-time layered-network epidemics model with time-varying transition rates and multiple resources. Automatica 2024, 159, 111303. [Google Scholar] [CrossRef]
- Klemm, S.; Ravera, L. On SIR-type epidemiological models and population heterogeneity effects. Physica A 2023, 624, 128928. [Google Scholar] [CrossRef]
- Melikechi, O.; Young, A.L.; Tang, T.; Bowman, T.; Dunson, D.; Johndrow, J. Limits of epidemic prediction using SIR models. Journal of Mathematical Biology 2022, 85, 36. [Google Scholar] [CrossRef] [PubMed]
- Machado, G.; Baxter, G.J. Effect of initial infection size on a network susceptible-infected-recovered model. Physical Review E 2022, 106, 014307. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.R. Research and Practice Analysis of Higher Vocational Colleges Facing the Experience and Dissemination of Regional Characteristic Tea Culture. Advances In Multimedia 2022, 2022, 2199788. [Google Scholar] [CrossRef]
- Jayatilaka, R.; Patel, R.; Brar, M.; Tang, Y.; Jisrawi, N.M.; Chishtie, F.; Drozd, J.; Valluri, S.R. A mathematical model of Covid-19 transmission. Materials Today-proceedings 2022, 54, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Palomo-Briones, G.A.; Siller, M.; Grignard, A. An agent-based model of the dual causality between individual and collective behaviors in an epidemic. Computers In Biology And Medicine 2022, 141, 104995. [Google Scholar] [CrossRef]
- Roman, H.E.; Croccolo, F. Spreading of Infections on Network Models: Percolation Clusters and Random Trees. Mathematics 2021, 9, 3054. [Google Scholar] [CrossRef]
- Kavitha, C.; Gowrisankar, A.; Banerjee, S. The second and third waves in India: when will the pandemic be culminated? European Physical Journal Plus 2021, 136, 596. [Google Scholar] [CrossRef]
- Di Crescenzo, A.; Gomez-Corral, A.; Taipe, D. A computational approach to extreme values and related hitting probabilities in level-dependent quasi-birth-death processes. Mathematics And Computers In Simulation 2025, 228, 211–224. [Google Scholar] [CrossRef]
- Yu, C.; He, J.M.; Ma, Q.T.; Liu, X.Y. Dynamic Evolution Model of Internet Financial Public Opinion. Information 2024, 15, 433. [Google Scholar] [CrossRef]
- Athapaththu, D.V.; Ambagaspitiya, T.D.; Chamberlain, A.; Demase, D.; Harasin, E.; Hicks, R.; Mcintosh, D.; Minute, G.; Petzold, S.; Tefft, L.; Chen, J.X. Physical Chemistry Lab for Data Analysis of Covid-19 Spreading Kinetics in Different Countries. Journal of Chemical Education 2024, 101, 2892–2898. [Google Scholar] [CrossRef]
- Song, Z.L.; Zhang, Z.; Lyu, F.; Bishop, M.; Liu, J.K.; Chi, Z.H. From Individual Motivation to Geospatial Epidemiology: A Novel Approach Using Fuzzy Cognitive Maps and Agent-Based Modeling for Large-Scale. Sustainability 2024, 16, 5036. [Google Scholar] [CrossRef]
- Li, G.J.; Chang, B.F.; Zhao, J.; Wang, J.Y.; He, F.; Wang, Y.H.; Xu, T.; Zhou, Z.G. VIVIAN: virtual simulation and visual analysis of epidemic spread data. Journal of Visualization 2024, 27, 677–694. [Google Scholar] [CrossRef]
- Agosto, A.; Cerchiello, P. A data-driven test approach to identify Covid-19 surge phases: an alert-warning tool. Statistics 2024, 58, 422–436. [Google Scholar] [CrossRef]
- Yoon, I.; Ahn, C.; Ahn, S.; Lee, B.; Lee, J.; Park, M. Enhancing indoor building occupant safety in the built environment: Assessing the validity of social force modeling for simulating physical. Developments In The Built Environment 2024, 17, 100336. [Google Scholar] [CrossRef]
- Yadav, S.K.; Khan, S.A.; Tiwari, M.; Kumar, A.; Kumar, V.; Akhter, Y. Taking cues from machine learning, compartmental and time series models for Sars-Cov-2 omicron infection in Indian provinces. Spatial And Spatio-temporal Epidemiology 2024, 48, 100634. [Google Scholar] [CrossRef]
- Finney, L.; Amundsen, D.E. Asymptotic analysis of periodic solutions of the seasonal SIR model. Physica D-nonlinear Phenomena 2024, 458, 133996. [Google Scholar] [CrossRef]
- Rocha, J.L.; Carvalho, S.; Coimbra, B. Probabilistic Procedures for SIR and SIS Epidemic Dynamics on Erdös-Rényi Contact Networks. Appliedmath 2023, 3, 828–850. [Google Scholar] [CrossRef]
- Ilic, B.; Salom, I.; Djordjevic, M.; Djordjevic, M. An analytical framework for understanding infection progression under social mitigation measures. Nonlinear Dynamics 2023, 111, 22033–22053. [Google Scholar] [CrossRef]
- Atienza-Diez, I.; Seoane, L.F. Long- and short-term effects of cross-immunity in epidemic dynamics. Chaos Solitons Fract. 2023, 174, 113800. [Google Scholar] [CrossRef]
- Prodanov, D. Computational aspects of the approximate analytic solutions of the SIR model: applications to modelling of Covid-19 outbreaks. Nonlinear Dynamics 2023, 111, 15613–15631. [Google Scholar] [CrossRef]
- Darvishi, H.; Darvishi, M.T. An Analytical Study on Two High-Order Hybrid Methods to Solve Systems of Nonlinear Equations. Journal of Mathematics 2023, 2023, 9917774. [Google Scholar] [CrossRef]
- Karaji, P.T.; Nyamoradi, N.; Ahmad, B. Stability and bifurcations of an SIR model with a nonlinear incidence rate. Mathematical Methods In The Applied Sciences 2023, 46, 10850–10866. [Google Scholar] [CrossRef]
- Chakir, Y. Global approximate solution of SIR epidemic model with constant vaccination strategy. Chaos Solitons Fract. 2023, 169, 113323. [Google Scholar] [CrossRef]
- Salimipour, A.; Mehraban, T.; Ghafour, H.S.; Arshad, N.I.; Ebadi, M.J. SIR model for the spread of Covid-19: A case study. Operations Research Perspectives 2023, 10, 100265. [Google Scholar] [CrossRef]
- Luangwilai, T.; Thammawichai, M. Optimal river flow management to reduce flood risks: A case study in northeastern Thailand. River Research And Applications 2023, 39, 255–265. [Google Scholar] [CrossRef]
- Prodanov, D. Asymptotic analysis of the SIR model and the Gompertz distribution. Journal of Computational And Applied Mathematics 2023, 422, 114901. [Google Scholar] [CrossRef]
- Qin, Y.P.; Wang, Z.; Zou, L. Analytical properties and solutions of a modified Lindemann mechanism with three reaction rate constants. Journal of Mathematical Chemistry 2023, 61, 389–401. [Google Scholar] [CrossRef]
- Ishfaq, U.; Khan, H.U.; Iqbal, S. Identifying the influential nodes in complex social networks using centrality-based approach. Journal of King Saud University-computer And Information Sciences 2022, 34, 9376–9392. [Google Scholar] [CrossRef]
- Gairat, A.; Shcherbakov, V. Discrete SIR model on a homogeneous tree and its continuous limit. Journal of Physics A 2022, 55, 434004. [Google Scholar] [CrossRef]
- Al-Obaidi, R.H.; Darvishi, M.T. Constructing a Class of Frozen Jacobian Multi-Step Iterative Solvers for Systems of Nonlinear Equations. Mathematics 2022, 10, 2952. [Google Scholar] [CrossRef]
- Öz, Y. Analytical investigation of compartmental models and measure for reactions of governments. European Physical Journal E 2022, 45, 68. [Google Scholar] [CrossRef] [PubMed]
- de Souza, D.B.; Araújo, H.A.; Duarte, G.C.; Gaffney, E.A.; Santos, F.A.N.; Raposo, E.P. Fock-space approach to stochastic susceptible-infected-recovered models. Physical Review E 2022, 106, 014136. [Google Scholar] [CrossRef] [PubMed]
- Kozyreff, G. Asymptotic solutions of the SIR and SEIR models well above the epidemic threshold. Ima Journal of Applied Mathematics 2022, 87, 521–536. [Google Scholar] [CrossRef]
- Tchoumi, S.Y.; Rwezaura, H.; Tchuenche, J.M. Dynamic of a two-strain Covid-19 model with vaccination. Results In Physics 2022, 39, 105777. [Google Scholar] [CrossRef]
- Schwarzendahl, F.J.; Grauer, J.; Liebchen, B.; Löwen, H. Mutation induced infection waves in diseases like Covid-19. Scientific Reports 2022, 12, 9641. [Google Scholar] [CrossRef]
- Gulec, F.; Atakan, B.; Dressler, F. Mobile human ad hoc networks: A communication engineering viewpoint on interhuman airborne pathogen transmission. Nano Communication Networks 2022, 32-33, 100410. [Google Scholar] [CrossRef]
- Turkyilmazoglu, M. A restricted epidemic SIR model with elementary solutions. Physica A 2022, 600, 127570. [Google Scholar] [CrossRef]
- Turkyilmazoglu, M. An extended epidemic model with vaccination: Weak-immune SIRV I. Physica A 2022, 598, 127429. [Google Scholar] [CrossRef]
- d’Andrea, V.; Gallotti, R.; Castaldo, N.; De Domenico, M. Individual risk perception and empirical social structures shape the dynamics of infectious disease outbreaks. Plos Computational Biology 2022, 18, e1009760. [Google Scholar] [CrossRef]
- Jiang, J.Y.; Zhou, Y.C.; Chen, X.S.; Jhou, Y.R.; Zhao, L.Q.; Liu, S.; Yang, P.C.; Ahmar, J.; Wang, W. COVID-19 Surveiller: toward a robust and effective pandemic surveillance system basedon social media mining. Philosophical Transactions of The Royal Society A-mathematical Physical 2022, 380, 20210125. [Google Scholar] [CrossRef]
- Uçar, D.; Çelik, E. Analysis of Covid 19 disease with SIR model and Taylor matrix method. Aims Mathematics 2022, 7, 11188–11200. [Google Scholar] [CrossRef]
- Berenbrink, P.; Cooper, C.; Gava, C.; Marzagao, D.K.; Mallmann-Trenn, F.; Radzik, T. On Early Extinction and the Effect of Travelling in the SIR model. Uncertainty In Artificial Intelligence, Vol 180 2022, 180, 159–169. [Google Scholar]
- Hussain, S.; Madi, E.N.; Khan, H.; Etemad, S.; Rezapour, S.; Sitthiwirattham, T.; Patanarapeelert, N. Investigation of the Stochastic Modeling of Covid-19 with Environmental Noise from the Analytical and Numerical Point of View. Mathematics 2021, 9, 3122. [Google Scholar] [CrossRef]
- Rusu, A.C.; Emonet, R.; Farrahi, K. Modelling digital and manual contact tracing for Covid-19. Are low uptakes and missed contacts deal-breakers? Plos One 2021, 16, e0259969. [Google Scholar] [CrossRef]
- Lee, K.; Parish, E.J. Parameterized neural ordinary differential equations: applications to computational physics problems. Proceedings of The Royal Society A-mathematical Physical And Engineering 2021, 477, 20210162. [Google Scholar] [CrossRef]
- Bärwolff, G. A Local and Time Resolution of the Covid-19 Propagation-A Two-Dimensional Approach for Germany Including Diffusion Phenomena to. Physics 2021, 3, 536–548. [Google Scholar] [CrossRef]
- Kartono, A.; Karimah, S.V.; Wahyudi, S.T.; Setiawan, A.A.; Sofian, I. Forecasting the Long-Term Trends of Coronavirus Disease 2019 (COVID-19) Epidemic Using the Susceptible-Infectious-Recovered (SIR) Model. Infectious Disease Reports 2021, 13, 668–684. [Google Scholar] [CrossRef]
- Hynd, R.; Ikpe, D.; Pendleton, T. Two critical times for the SIR model. Journal of Mathematical Analysis And Applications 2022, 505, 125507. [Google Scholar] [CrossRef]
- Zhou, Y.C.; Jiang, J.Y.; Chen, X.S.; Wang, W. #StayHome or #Marathon? Social Media Enhanced Pandemic Surveillance on Spatial-temporal Dynamic Graphs. Proceedings of The 30th Acm International Conference On Information 2021, pp. 2738–2748.
- Schüttler, J.; Schlickeiser, R.; Schlickeiser, F.; Kröger, M. Covid-19 predictions using a Gauss model, based on data from April 2. Physics 2020, 2, 197–202. [Google Scholar] [CrossRef]
- Kröger, M.; Turkyilmazoglu, M.; Schlickeiser, R. Explicit formulae for the peak time of an epidemic from the SIR model. Which approximant to use? Physica D 2021, 425, 132981. [Google Scholar] [CrossRef]
- Lambert, J.H. Observations variae in Mathesin Puram, Acta Helvitica. Phys. Math. Anatom. Botan Med. 1758, 3, 128–168. [Google Scholar]
- Kröger, M.; Schlickeiser, R. Analytical solution of the SIR-model for the temporal evolution of epidemics. Part A: Time-independent reproduction factor. J. Phys. A: Math. Theor. 2020, 53, 505601. [Google Scholar] [CrossRef]
- Haas, F.; Kröger, M.; Schlickeiser, R. Multi-Hamiltonian structure of the epidemics model accounting for vaccinations and a suitable test for the accuracy of its numerical solvers. J. Phys. A 2022, 55, 225206. [Google Scholar] [CrossRef]
- Weisstein, E. The CRC Encyclopedia of Mathematics, Third Edition; Chapman and Hall/CRC Press, Boca Raton, Florida, United States, 2009.
- Beyer, W.H. CRC Standard Mathematical Tables, 28th ed.; CRC Press, Boca Raton, Florida, United States, 1987; p. 455.
- Shampine, L.F.; Reichelt, M.W. The MATLABODE suite. SIAM J. Sci. Comput. 1997, 18, 1–22. [Google Scholar] [CrossRef]
- Bender, C.; Ghosh, A.; Vakili, H.; Ghosh, P.; Ghosh, A.W. An effective drift-diffusion model for pandemic propagation and uncertainty prediction. Biophysical Reports 2024, 4, 100182. [Google Scholar] [CrossRef]
- Jang, G.H.; Kim, S.J.; Lee, M.J.; Son, S.W. Effectiveness of vaccination and quarantine policies to curb the spread of Covid-19. Physica A 2024, 637, 129580. [Google Scholar] [CrossRef]
- Gao, W.H.; Wang, Y.; Cao, J.D.; Liu, Y. Final epidemic size and critical times for susceptible-infectious-recovered models with a generalized contact rate. Chaos 2024, 34, 013152. [Google Scholar] [CrossRef]
- Dobie, A.P.; Bayrakal, A.; Or, M.E.; Bilge, A.H. Dynamics of Feline Coronavirus and FIP: A Compartmental Modeling Approach. Veterinary Medicine International 2023, 2023, 2721907. [Google Scholar] [CrossRef]
- Shirazian, M. A new acceleration of variational iteration method for initial value problems. Mathematics And Computers In Simulation 2023, 214, 246–259. [Google Scholar] [CrossRef]
- Turkyilmazoglu, M. A highly accurate peak time formula of epidemic outbreak from the SIR model. Chinese Journal of Physics 2023, 84, 39–50. [Google Scholar] [CrossRef]
- Alshammari, F.S. Analysis of SIRV I model with time dependent coefficients and the effect of vaccination on the transmission rate and Covid- 19 epidemic waves. Infectious Disease Modelling 2023, 8, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Dimou, A.; Maragakis, M.; Argyrakis, P. A network SIRX model for the spreading of Covid-19. Physica A 2022, 590, 126746. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.B.; Wu, H. There exists the ’smartest’ movement rate to control the epidemic rather than ’city lockdown’? Applied Mathematical Modelling 2022, 106, 696–714. [Google Scholar] [CrossRef] [PubMed]
- Gölgeli, M. The contagion dynamics of vaccine skepticism. Hacettepe Journal of Mathematics And Statistics 2022, 51, 1697–1709. [Google Scholar] [CrossRef]
- Wang, S.F.; Ma, Z.H.; Li, X.H.; Qi, T. A generalized delay-induced SIRS epidemic model with relapse. Aims Mathematics 2022, 7, 6600–6618. [Google Scholar] [CrossRef]
- Harko, T.; Lobo, F.S.N.; Mak, M.K. Exact analytical solutions of the susceptible-infected-recovered (SIR) epidemic model and of the SIR model with equal death and birth rates. Appl. Math. Comput. 2014, 236, 184–194. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).