Submitted:
27 November 2024
Posted:
29 November 2024
You are already at the latest version
Abstract
Keywords:
1. Soil Salinisation Is a Major Problem Under Climate Change
1.1. Origins and Consequences of Soil Salinisation
1.2. Why the Olive Tree?
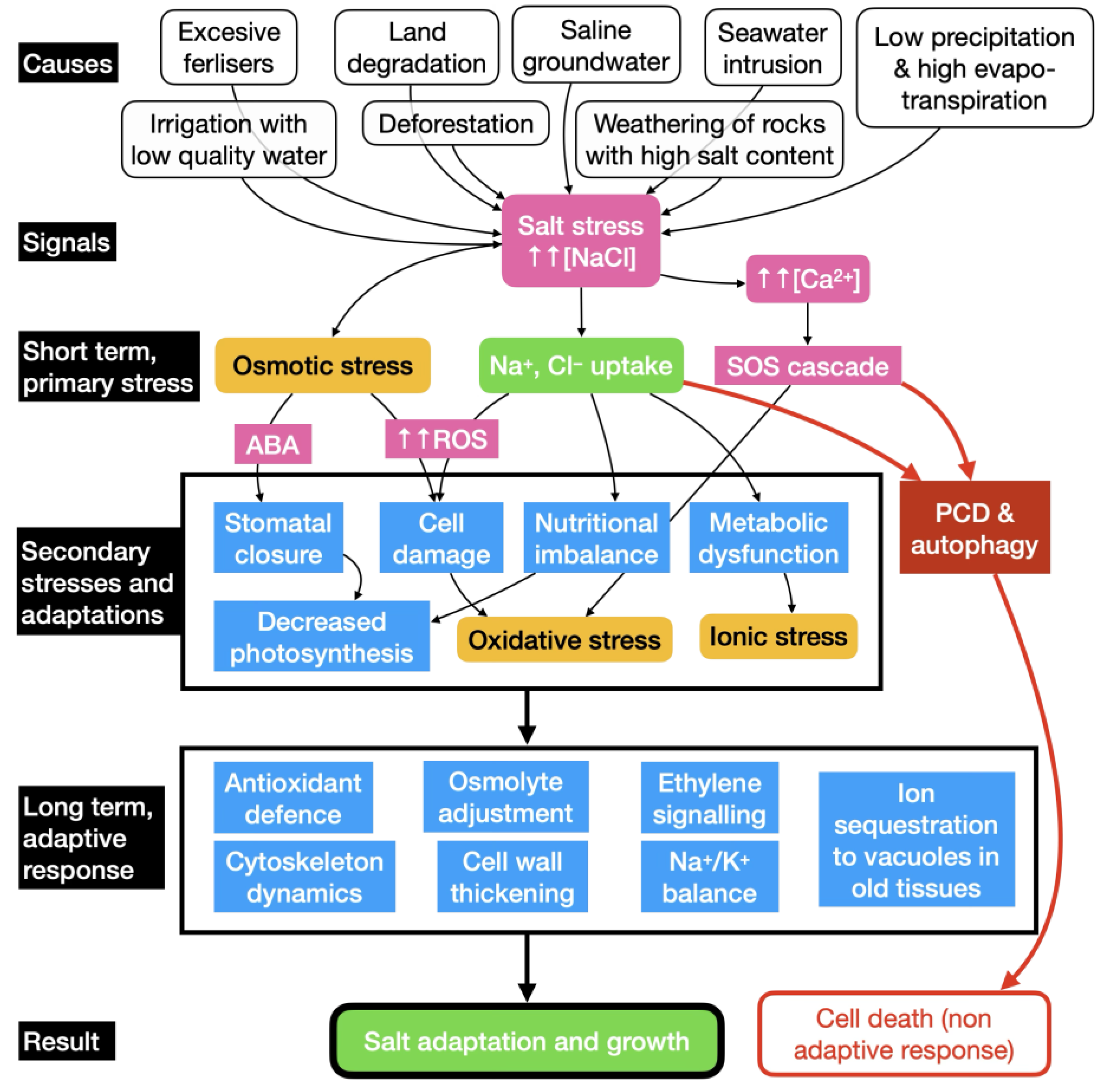
2. Plant Management of Soil Salinisation
2.1. Overview of Plant Responses to Salt Exposure
2.2. Biochemical Indicators of Salt Adaptation
2.3. Adaptive Signalling to Salt Stress
2.3.1. Hormonal Signalling
2.3.2. Post-Translational Modifications
2.3.3. Signalling Crosstalk
2.4. Cell Wall Thickening and Leaf Changes Under Salt Stress
2.5. Avoiding Ion Toxicity
2.6. Programmed Cell Death and Autophagy Under Salt Stress Conditions
2.7. Computer Learning as an Integrative Approach to Study Salt Stress
3. Olive Tree Management of Soil Salinisation
3.1. Ion Exclusion and Retention in Olive Tree
3.2. Biochemical Adaptations to Salt in Olive Cultivars
3.3. Salt Tolerance of Olive Tree is Cultivar-Dependent
4. Studies About Salt Stress Genomics in Olive Tree
4.1. More and More Genome Sequences of Olive Cultivars Are Available
4.2. Transcriptional Response to Salt Stress in Olive Tree
5. Microbiota Changes in Salinised Soils of Olive Groves
5.1. Distribution of Plant-Associated Microorganisms
5.2. The Olive Grove Microbiota
5.3. Decrease of Microbiota Biodiversity due to Agricultural Practices
5.4. The Olive Tree Microbiota Changes Under Salt Stress
5.5. Soil Amendments and Salt Stress
6. Bioinformatics Studies in Olive Tree Concerning Salt Stress
6.1. From Olive Tree Transcriptome to OliveAtlas
6.2. Machine Learning to Study the Olive Tree
7. Conclusions: Stepping Towards a 'Smart Oliviculture'
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Daliakopoulos, I.N.; Tsanis, I.K.; Koutroulis, A.; Kourgialas, N.N.; Varouchakis, A.E.; Karatzas, G.P.; Ritsema, C.J. The Threat of Soil Salinity: A European Scale Review. Sci. Total Environ. 2016, 573, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Hassani, A.; Azapagic, A.; Shokri, N. Global Predictions of Primary Soil Salinization under Changing Climate in the 21st Century. Nat. Commun. 2021, 12, 6663. [Google Scholar] [CrossRef] [PubMed]
- Calvin, K.; Dasgupta, D.; Krinner, G.; Mukherji, A.; Thorne, P.W.; Trisos, C.; Romero, J.; Aldunce, P.; Barrett, K.; Blanco, G.; et al. IPCC, 2023: Climate Change 2023: Synthesis Report. Contribution of Working Groups I, II and III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change [Core Writing Team, H. Lee and J. Romero (Eds.)]. IPCC, Geneva, Switzerland.; First.; Intergovernmental Panel on Climate Change (IPCC), 2023;
- Shahid, S.A.; Zaman, M.; Heng, L. Introduction to Soil Salinity, Sodicity and Diagnostics Techniques. In Guideline for Salinity Assessment, Mitigation and Adaptation Using Nuclear and Related Techniques; Springer International Publishing: Cham, 2018; ISBN 978-3-319-96189-7. [Google Scholar]
- Whittle, A.; Barnett, R.L.; Charman, D.J.; Gallego-Sala, A.V. Low-Salinity Transitions Drive Abrupt Microbial Response to Sea-Level Change. Ecol. Lett. 2022, 25, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, P.; Kumar, R. Soil Salinity: A Serious Environmental Issue and Plant Growth Promoting Bacteria as One of the Tools for Its Alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef]
- Singh, A. Soil Salinity: A Global Threat to Sustainable Development. Soil Use Manag. 2022, 38, 39–67. [Google Scholar] [CrossRef]
- Stavi, I.; Thevs, N.; Priori, S. Soil Salinity and Sodicity in Drylands: A Review of Causes, Effects, Monitoring, and Restoration Measures. Front. Environ. Sci. 2021, 9, 712831. [Google Scholar] [CrossRef]
- Majeed, A.; Siyyar, S. Salinity Stress Management in Field Crops: An Overview of the Agronomic Approaches. In Plant Ecophysiology and Adaptation under Climate Change: Mechanisms and Perspectives II.; Hasanuzzaman, M., Ed.; Springer Singapore: Singapore, 2020; ISBN 9789811521713. [Google Scholar]
- Romero-Trigueros, C.; Vivaldi, G.A.; Nicolás, E.N.; Paduano, A.; Salcedo, F.P.; Camposeo, S. Ripening Indices, Olive Yield and Oil Quality in Response to Irrigation With Saline Reclaimed Water and Deficit Strategies. Front. Plant Sci. 2019, 10, 1243. [Google Scholar] [CrossRef]
- Meena, M.D.; Yadav, R.K.; Narjary, B.; Yadav, G.; Jat, H.S.; Sheoran, P.; Meena, M.K.; Antil, R.S.; Meena, B.L.; Singh, H.V.; et al. Municipal Solid Waste (MSW): Strategies to Improve Salt Affected Soil Sustainability: A Review. Waste Manag. 2019, 84, 38–53. [Google Scholar] [CrossRef]
- Munns, R.; Gilliham, M. Salinity Tolerance of Crops – What Is the Cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef]
- Chele, K.H.; Tinte, M.M.; Piater, L.A.; Dubery, I.A.; Tugizimana, F. Soil Salinity, a Serious Environmental Issue and Plant Responses: A Metabolomics Perspective. Metabolites 2021, 11, 724. [Google Scholar] [CrossRef]
- Atta, K.; Mondal, S.; Gorai, S.; Singh, A.P.; Kumari, A.; Ghosh, T.; Roy, A.; Hembram, S.; Gaikwad, D.J.; Mondal, S.; et al. Impacts of Salinity Stress on Crop Plants: Improving Salt Tolerance through Genetic and Molecular Dissection. Front. Plant Sci. 2023, 14, 1241736. [Google Scholar] [CrossRef]
- El Yamani, M.; Cordovilla, M.D.P. Tolerance Mechanisms of Olive Tree (Olea europaea) Under Saline Conditions. Plants 2024, 13, 2094. [Google Scholar] [CrossRef] [PubMed]
- Fraga, H.; Moriondo, M.; Leolini, L.; Santos, J.A. Mediterranean Olive Orchards under Climate Change: A Review of Future Impacts and Adaptation Strategies. Agronomy 2020, 11, 56. [Google Scholar] [CrossRef]
- Dini, I.; Laneri, S. Spices, Condiments, Extra Virgin Olive Oil and Aromas as Not Only Flavorings, but Precious Allies for Our Wellbeing. Antioxidants 2021, 10, 868. [Google Scholar] [CrossRef]
- Unver, T.; Wu, Z.; Sterck, L.; Turktas, M.; Lohaus, R.; Li, Z.; Yang, M.; He, L.; Deng, T.; Escalante, F.J.; et al. Genome of Wild Olive and the Evolution of Oil Biosynthesis. Proc. Natl. Acad. Sci. 2017, 114, E9413–E9422. [Google Scholar] [CrossRef]
- Maestri, D.; Barrionuevo, D.; Bodoira, R.; Zafra, A.; Jiménez-López, J.; Alché, J. de D. Nutritional Profile and Nutraceutical Components of Olive (Olea europaea L.) Seeds. J. Food Sci. Technol. 2019, 56, 4359–4370. [Google Scholar] [CrossRef]
- Georgiadou, E.C.; Koubouris, G.; Goulas, V.; Sergentani, C.; Nikoloudakis, N.; Manganaris, G.A.; Kalaitzis, P.; Fotopoulos, V. Genotype-dependent Regulation of Vitamin E Biosynthesis in Olive Fruits as Revealed through Metabolic and Transcriptional Profiles. Plant Biol. 2019, 21, 604–614. [Google Scholar] [CrossRef]
- Bruno, L.; Picardi, E.; Pacenza, M.; Chiappetta, A.; Muto, A.; Gagliardi, O.; Muzzalupo, I.; Pesole, G.; Bitonti, M.B. Changes in Gene Expression and Metabolic Profile of Drupes of Olea europaea L. Cv Carolea in Relation to Maturation Stage and Cultivation Area. BMC Plant Biol. 2019, 19, 428. [Google Scholar] [CrossRef]
- Mougiou, N.; Trikka, F.; Trantas, E.; Ververidis, F.; Makris, A.; Argiriou, A.; Vlachonasios, K.E. Expression of Hydroxytyrosol and Oleuropein Biosynthetic Genes Are Correlated with Metabolite Accumulation during Fruit Development in Olive, Olea Europaea, Cv. Koroneiki. Plant Physiol. Biochem. 2018, 128, 41–49. [Google Scholar] [CrossRef]
- Iaria, D.L.; Chiappetta, A.; Muzzalupo, I. A De Novo Transcriptomic Approach to Identify Flavonoids and Anthocyanins “Switch-Off” in Olive (Olea europaea L.) Drupes at Different Stages of Maturation. Front. Plant Sci. 2016, 6, 1246. [Google Scholar] [CrossRef]
- Parvini, F.; Sicardo, M.D.; Hosseini-Mazinani, M.; Martínez-Rivas, J.M.; Hernández, M.L. Transcriptional Analysis of Stearoyl-Acyl Carrier Protein Desaturase Genes from Olive (Olea europaea ) in Relation to the Oleic Acid Content of the Virgin Olive Oil. J. Agric. Food Chem. 2016, 64, 7770–7781. [Google Scholar] [CrossRef] [PubMed]
- Yanik, H.; Turktas, M.; Dundar, E.; Hernandez, P.; Dorado, G.; Unver, T. Genome-Wide Identification of Alternate Bearing-Associated microRNAs (miRNAs) in Olive (Olea europaea L.). BMC Plant Biol. 2013, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- García-López, M.C.; Vidoy, I.; Jiménez-Ruiz, J.; Muñoz-Mérida, A.; Fernández-Ocaña, A.; de la Rosa, R.; Barroso, J.B.; Navarro, F.; Trelles, O.; Beuzón, C.R.; et al. Genetic Changes Involved in the Juvenile-to-Adult Transition in the Shoot Apex of Olea europaea L. Occur Years before the First Flowering. Tree Genet. Genomes 2014, 10, 585–603. [Google Scholar] [CrossRef]
- Jiménez-Ruiz, J.; García-López, M.C.; Vidoy, I.; de la O Leyva-Pérez, M.; Fernández-Ocaña, A.; Barroso, J.B.; Barceló, A.; Beuzón, C.R.; de la Rosa, R.; Luque, F. Transcriptional Analysis of Adult Cutting and Juvenile Seedling Olive Roots. Tree Genet. Genomes 2015, 11, 77. [Google Scholar] [CrossRef]
- Carmona, R.; Zafra, A.; Seoane, P.; Castro, A.J.; Guerrero-Fernández, D.; Castillo-Castillo, T.; Medina-García, A.; Cánovas, F.M.; Aldana-Montes, J.F.; Navas-Delgado, I.; et al. ReprOlive: A Database with Linked Data for the Olive Tree (Olea europaea L.) Reproductive Transcriptome. Front. Plant Sci. 2015, 6, 625. [Google Scholar] [CrossRef]
- Rejón, J.; Delalande, F.; Schaeffer-Reiss, C.; Alché, J.; Rodríguez-García, M.; Van Dorsselaer, A.; Castro, A. The Pollen Coat Proteome: At the Cutting Edge of Plant Reproduction. Proteomes 2016, 4, 5. [Google Scholar] [CrossRef]
- Zafra, A.; Carmona, R.; Traverso, J.A.; Hancock, J.T.; Goldman, M.H.S.; Claros, M.G.; Hiscock, S.J.; Alche, J.D. Identification and Functional Annotation of Genes Differentially Expressed in the Reproductive Tissues of the Olive Tree (Olea europaea L.) through the Generation of Subtractive Libraries. Front. Plant Sci. 2017, 8, 1576. [Google Scholar] [CrossRef]
- Farinelli, D.; Breton, C.; Koubouris, G.; Famiani, F.; Villemur, P.; Bervillé, A. Reply to Saumitou-Laprade et al. (2017) “Controlling for Genetic Identity of Varieties, Pollen Contamination and Stigma Receptivity Is Essential to Characterize the Self-Incompatibility System of Olea europaea L.”. Evol. Appl. 2018, 11, 1465–1470. [Google Scholar] [CrossRef]
- Alagna, F.; Caceres, M.E.; Pandolfi, S.; Collani, S.; Mousavi, S.; Mariotti, R.; Cultrera, N.G.M.; Baldoni, L.; Barcaccia, G. The Paradox of Self-Fertile Varieties in the Context of Self-Incompatible Genotypes in Olive. Front. Plant Sci. 2019, 10, 725. [Google Scholar] [CrossRef]
- Bullones, A.; Castro, A.J.; Lima-Cabello, E.; Fernandez-Pozo, N.; Bautista, R.; Alché, J.D.D.; Claros, M.G. Transcriptomic Insight into the Pollen Tube Growth of Olea europaea L. subsp. europaea Reveals Reprogramming and Pollen-Specific Genes Including New Transcription Factors. Plants 2023, 12, 2894. [Google Scholar] [CrossRef]
- Gros-Balthazard, M.; Besnard, G.; Sarah, G.; Holtz, Y.; Leclercq, J.; Santoni, S.; Wegmann, D.; Glémin, S.; Khadari, B. Evolutionary Transcriptomics Reveals the Origins of Olives and the Genomic Changes Associated with Their Domestication. Plant J. 2019, 100, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Castro, A.J.; de Dios Alché, J.; Cuevas, J.; Romero, P.J.; Alché, V.; Rodríguez-García, M.I. Pollen from Different Olive Tree Cultivars Contains Varying Amounts of the Major Allergen Ole e 1. Int. Arch. Allergy Immunol. 2003, 131, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Alché, J.D.; Castro, A.J.; Jiménez-López, J.C.; Morales, S.; Zafra, A.; Hamman-Khalifa, A.M.; Rodríguez-García, M.I. Differential Characteristics of Olive Pollen from Different Cultivars: Biological and Clinical Implications. J. Investig. Allergol. Clin. Immunol. 2007, 17 Suppl 1, 17–23. [Google Scholar]
- Hamman-Khalifa, A.; Castro, A.; Jiménez-López, J.; Rodríguez-García, M.; Alché, J. Olive Cultivar Origin Is a Major Cause of Polymorphism for Ole e 1 Pollen Allergen. BMC Plant Biol. 2008, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Grasso, F.; Coppola, M.; Carbone, F.; Baldoni, L.; Alagna, F.; Perrotta, G.; Pérez-Pulido, A.J.; Garonna, A.; Facella, P.; Daddiego, L.; et al. The Transcriptional Response to the Olive Fruit Fly (Bactrocera oleae) Reveals Extended Differences between Tolerant and Susceptible Olive (Olea europaea L.) Varieties. PLOS ONE 2017, 12, e0183050. [Google Scholar] [CrossRef]
- León, L.; De La Rosa, R.; Arriaza, M. Prioritization of Olive Breeding Objectives in Spain: Analysis of a Producers and Researchers Survey. Span. J. Agric. Res. 2021, 19, e0701. [Google Scholar] [CrossRef]
- Gucci, R.; Tattini, M. Salinity Tolerance in Olive. In Horticultural Reviews; Janick, J., Ed.; John Wiley & Sons, Inc.: Oxford, UK, 2010; ISBN 978-0-470-65066-0. [Google Scholar]
- Lavee, S. Evaluation of the Need and Present Potential of Olive Breeding Indicating the Nature of the Available Genetic Resources Involved. Sci. Hortic. 2013, 161, 333–339. [Google Scholar] [CrossRef]
- Hualpa-Ramirez, E.; Carrasco-Lozano, E.C.; Madrid-Espinoza, J.; Tejos, R.; Ruiz-Lara, S.; Stange, C.; Norambuena, L. Stress Salinity in Plants: New Strategies to Cope with in the Foreseeable Scenario. Plant Physiol. Biochem. 2024, 208, 108507. [Google Scholar] [CrossRef]
- Romero-Aranda, R.; Moya, J.L.; Tadeo, F.R.; Legaz, F.; Primo-Millo, E.; Talon, M. Physiological and Anatomical Disturbances Induced by Chloride Salts in Sensitive and Tolerant Citrus: Beneficial and Detrimental Effects of Cations. Plant Cell Environ. 1998, 21, 1243–1253. [Google Scholar] [CrossRef]
- Romero-Aranda, R.; Soria, T.; Cuartero, J. Tomato Plant-Water Uptake and Plant-Water Relationships under Saline Growth Conditions. Plant Sci. 2001, 160, 265–272. [Google Scholar] [CrossRef]
- Muchate, N.S.; Nikalje, G.C.; Rajurkar, N.S.; Suprasanna, P.; Nikam, T.D. Plant Salt Stress: Adaptive Responses, Tolerance Mechanism and Bioengineering for Salt Tolerance. Bot. Rev. 2016, 82, 371–406. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Romero-Aranda, M.R.; González-Fernández, P.; Pérez-Tienda, J.R.; López-Diaz, M.R.; Espinosa, J.; Granum, E.; Traverso, J.Á.; Pineda, B.; Garcia-Sogo, B.; Moreno, V.; et al. Na+ Transporter HKT1;2 Reduces Flower Na+ Content and Considerably Mitigates the Decline in Tomato Fruit Yields under Saline Conditions. Plant Physiol. Biochem. 2020, 154, 341–352. [Google Scholar] [CrossRef]
- Niu, G.; Rodriguez, D.S.; Starman, T. Response of Bedding Plants to Saline Water Irrigation. HortScience 2010, 45, 628–636. [Google Scholar] [CrossRef]
- Sánchez-Blanco, M.J.; Álvarez, S.; Ortuño, M.F.; Ruiz-Sánchez, M.C. Root System Response to Drought and Salinity: Root Distribution and Water Transport. In Root Engineering; Morte, A., Varma, A., Eds.; Soil Biology; Springer Berlin Heidelberg: Berlin, Heidelberg, 2014; ISBN 978-3-642-54275-6. [Google Scholar]
- Plant Roots. The Hidden Half; Waisel, Y., Eshel, A., Beeckman, T., Kafkafi, U., Eds.; 3rd ed.; CRC Press: Boca Ratón, 2002; ISBN 978-0-203-90942-3. [Google Scholar]
- Tu, T.Q.; Vaciaxa, P.; Lo, T.T.M.; Nguyen, N.H.; Pham, N.T.T.; Nguyen, Q.H.; Do, P.T.; Nguyen, L.T.N.; Nguyen, Y.T.H.; Chu, M.H. GmDREB6, a Soybean Transcription Factor, Notably Affects the Transcription of the NtP5CS and NtCLC Genes in Transgenic Tobacco under Salt Stress Conditions. Saudi J. Biol. Sci. 2021, 28, 7175–7181. [Google Scholar] [CrossRef]
- Das, K.; Roychoudhury, A. Reactive Oxygen Species (ROS) and Response of Antioxidants as ROS-Scavengers during Environmental Stress in Plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- Singh, A.; Roychoudhury, A. Gene Regulation at Transcriptional and Post-transcriptional Levels to Combat Salt Stress in Plants. Physiol. Plant. 2021, 173, 1556–1572. [Google Scholar] [CrossRef]
- Balasubramaniam, T.; Shen, G.; Esmaeili, N.; Zhang, H. Plants’ Response Mechanisms to Salinity Stress. Plants 2023, 12, 2253. [Google Scholar] [CrossRef]
- Mansour, M.M.F.; Hassan, F.A.S. How Salt Stress-Responsive Proteins Regulate Plant Adaptation to Saline Conditions. Plant Mol. Biol. 2022, 108, 175–224. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.-K.; Bohnert, H.J. Plant Celular and Molecular Responses to High Salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef]
- Bruetschy, C. The EU Regulatory Framework on Genetically Modified Organisms (GMOs). Transgenic Res. 2019, 28, 169–174. [Google Scholar] [CrossRef]
- Soltabayeva, A.; Ongaltay, A.; Omondi, J.O.; Srivastava, S. Morphological, Physiological and Molecular Markers for Salt-Stressed Plants. Plants 2021, 10, 243. [Google Scholar] [CrossRef]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Afzal, M.Z.; Jia, Q.; Ibrahim, A.K.; Niyitanga, S.; Zhang, L. Mechanisms and Signaling Pathways of Salt Tolerance in Crops: Understanding from the Transgenic Plants. Trop. Plant Biol. 2020, 13, 297–320. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Zhou, H.; Shi, H.; Yang, Y.; Feng, X.; Chen, X.; Xiao, F.; Lin, H.; Guo, Y. Insights into Plant Salt Stress Signaling and Tolerance. J. Genet. Genomics Yi Chuan Xue Bao 2024, 51, 16–34. [Google Scholar] [CrossRef]
- Najar, M.A. Updates on Protein Post-Translational Modifications for Modulating Response to Salinity. In Genetics of Salt Tolerance in Plants; Ganie, S.A., Wani, S.H., Eds.; CABI: GB, 2024; ISBN 978-1-80062-301-9. [Google Scholar]
- Mata-Pérez, C.; Sánchez-Vicente, I.; Arteaga, N.; Gómez-Jiménez, S.; Fuentes-Terrón, A.; Oulebsir, C.S.; Calvo-Polanco, M.; Oliver, C.; Lorenzo, Ó. Functions of Nitric Oxide-Mediated Post-Translational Modifications under Abiotic Stress. Front. Plant Sci. 2023, 14, 1158184. [Google Scholar] [CrossRef]
- Benjamin, J.J.; Miras-Moreno, B.; Araniti, F.; Salehi, H.; Bernardo, L.; Parida, A.; Lucini, L. Proteomics Revealed Distinct Responses to Salinity between the Halophytes Suaeda maritima (L.) Dumort and Salicornia brachiata (Roxb). Plants 2020, 9, 227. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Nafees, M.; Chen, J.; Darras, A.; Ferrante, A.; Hancock, J.T.; Ashraf, M.; Zaid, A.; Latif, N.; Corpas, F.J.; et al. Chemical Priming Enhances Plant Tolerance to Salt Stress. Front. Plant Sci. 2022, 13, 946922. [Google Scholar] [CrossRef]
- Tang, R.-J.; Wang, C.; Li, K.; Luan, S. The CBL-CIPK Calcium Signaling Network: Unified Paradigm from 20 Years of Discoveries. Trends Plant Sci. 2020, 25, 604–617. [Google Scholar] [CrossRef]
- Ismail, A.; Takeda, S.; Nick, P. Life and Death under Salt Stress: Same Players, Different Timing? J. Exp. Bot. 2014, 65, 2963–2979. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Raja, V.; Majeed, U.; Kang, H.; Andrabi, K.I.; John, R. Abiotic Stress: Interplay between ROS, Hormones and MAPKs. Environ. Exp. Bot. 2017, 137, 142–157. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.; Gaurav, A.K.; Srivastava, S.; Verma, J.P. Plant Growth-Promoting Bacteria: Biological Tools for the Mitigation of Salinity Stress in Plants. Front. Microbiol. 2020, 11, 1216. [Google Scholar] [CrossRef]
- Dabravolski, S.A.; Isayenkov, S.V. The Regulation of Plant Cell Wall Organisation under Salt Stress. Front. Plant Sci. 2023, 14, 1118313. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, W.; Long, S.; Zhao, C. Maintenance of Cell Wall Integrity under High Salinity. Int. J. Mol. Sci. 2021, 22, 3260. [Google Scholar] [CrossRef]
- Lei, X.; Liu, Z.; Xie, Q.; Fang, J.; Wang, C.; Li, J.; Wang, C.; Gao, C. Construction of Two Regulatory Networks Related to Salt Stress and Lignocellulosic Synthesis under Salt Stress Based on a Populus davidiana × P. bolleana Transcriptome Analysis. Plant Mol. Biol. 2022, 109, 689–702. [Google Scholar] [CrossRef]
- Venkataraman, G.; Shabala, S.; Véry, A.-A.; Hariharan, G.N.; Somasundaram, S.; Pulipati, S.; Sellamuthu, G.; Harikrishnan, M.; Kumari, K.; Shabala, L.; et al. To Exclude or to Accumulate? Revealing the Role of the Sodium HKT1;5 Transporter in Plant Adaptive Responses to Varying Soil Salinity. Plant Physiol. Biochem. 2021, 169, 333–342. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity Tolerance in Halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef]
- Cheeseman, J.M. The Evolution of Halophytes, Glycophytes and Crops, and Its Implications for Food Security under Saline Conditions. New Phytol. 2015, 206, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, P.M. Sodium (Na+) Homeostasis and Salt Tolerance of Plants. Environ. Exp. Bot. 2013, 92, 19–31. [Google Scholar] [CrossRef]
- Hussain, S.; Zhang, J.; Zhong, C.; Zhu, L.; Cao, X.; Yu, S.; Allen Bohr, J.; Hu, J.; Jin, Q. Effects of Salt Stress on Rice Growth, Development Characteristics, and the Regulating Ways: A Review. J. Integr. Agric. 2017, 16, 2357–2374. [Google Scholar] [CrossRef]
- Kronzucker, H.J.; Coskun, D.; Schulze, L.M.; Wong, J.R.; Britto, D.T. Sodium as Nutrient and Toxicant. Plant Soil 2013, 369, 1–23. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Gilliham, M.; Flowers, T.J.; Colmer, T.D. Tissue Tolerance: An Essential but Elusive Trait for Salt-Tolerant Crops. Funct. Plant Biol. 2016, 43, 1103. [Google Scholar] [CrossRef]
- Tester, M. Na+ Tolerance and Na+ Transport in Higher Plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef]
- Brumós, J.; Colmenero-Flores, J.M.; Conesa, A.; Izquierdo, P.; Sánchez, G.; Iglesias, D.J.; López-Climent, M.F.; Gómez-Cadenas, A.; Talón, M. Membrane Transporters and Carbon Metabolism Implicated in Chloride Homeostasis Differentiate Salt Stress Responses in Tolerant and Sensitive Citrus Rootstocks. Funct. Integr. Genomics 2009, 9, 293–309. [Google Scholar] [CrossRef]
- Brumós, J.; Talón, M.; Bouhlal, R.; Colmenero-Flores, J.M. Cl- Homeostasis in Includer and Excluder Citrus Rootstocks: Transport Mechanisms and Identification of Candidate Genes. Plant Cell Environ. 2010, 33, 2012–2027. [Google Scholar] [CrossRef]
- Li, B.; Tester, M.; Gilliham, M. Chloride on the Move. Trends Plant Sci. 2017, 22, 236–248. [Google Scholar] [CrossRef]
- Celis, N.; Suarez, D.L.; Wu, L.; Li, R.; Arpaia, M.L.; Mauk, P. Salt Tolerance and Growth of 13 Avocado Rootstocks Related Best to Chloride Uptake. HortScience 2018, 53, 1737–1745. [Google Scholar] [CrossRef]
- Colmenero-Flores, J.M.; Franco-Navarro, J.D.; Cubero-Font, P.; Peinado-Torrubia, P.; Rosales, M.A. Chloride as a Beneficial Macronutrient in Higher Plants: New Roles and Regulation. Int. J. Mol. Sci. 2019, 20, 4686. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sandhu, N.; Kumar, P.; Pruthi, G.; Singh, J.; Kaur, S.; Chhuneja, P. Genome-Wide Identification and in Silico Analysis of NPF, NRT2, CLC and SLAC1/SLAH Nitrate Transporters in Hexaploid Wheat (Triticum aestivum). Sci. Rep. 2022, 12, 11227. [Google Scholar] [CrossRef] [PubMed]
- Rajappa, S.; Krishnamurthy, P.; Huang, H.; Yu, D.; Friml, J.; Xu, J.; Kumar, P.P. The Translocation of a Chloride Channel from the Golgi to the Plasma Membrane Helps Plants Adapt to Salt Stress. Nat. Commun. 2024, 15, 3978. [Google Scholar] [CrossRef] [PubMed]
- Wleklik, K.; Borek, S. Vacuolar Processing Enzymes in Plant Programmed Cell Death and Autophagy. Int. J. Mol. Sci. 2023, 24, 1198. [Google Scholar] [CrossRef] [PubMed]
- Aina, O.; Bakare, O.O.; Fadaka, A.O.; Keyster, M.; Klein, A. Plant Biomarkers as Early Detection Tools in Stress Management in Food Crops: A Review. Planta 2024, 259, 60. [Google Scholar] [CrossRef]
- Mani, B.; Kaur, I.; Dhingra, Y.; Saxena, V.; Krishna, G.K.; Kumar, R.; Chinnusamy, V.; Agarwal, M.; Katiyar-Agarwal, S. Tetraspanin 5 Orchestrates Resilience to Salt Stress through the Regulation of Ion and Reactive Oxygen Species Homeostasis in Rice. Plant Biotechnol. J. 2024, pbi.14476. [CrossRef]
- Kotula, L.; Garcia Caparros, P.; Zörb, C.; Colmer, T.D.; Flowers, T.J. Improving Crop Salt Tolerance Using Transgenic Approaches: An Update and Physiological Analysis. Plant Cell Environ. 2020, 43, 2932–2956. [Google Scholar] [CrossRef]
- Stokstad, E. European Parliament Votes to Ease Regulation of Gene-Edited Crops. Science 2024. [Google Scholar] [CrossRef]
- Puchta, H. Regulation of Gene-Edited Plants in Europe: From the Valley of Tears into the Shining Sun? aBIOTECH 2023, 5, 231–238. [Google Scholar] [CrossRef]
- Affenzeller, M.J.; Darehshouri, A.; Andosch, A.; Lutz, C.; Lutz-Meindl, U. Salt Stress-Induced Cell Death in the Unicellular Green Alga Micrasterias denticulata. J. Exp. Bot. 2009, 60, 939–954. [Google Scholar] [CrossRef]
- Joseph, B.; Jini, D. Salinity Induced Programmed Cell Death in Plants: Challenges and Opportunities for Salt-Tolerant Plants. J. Plant Sci. 2010, 5, 376–390. [Google Scholar] [CrossRef]
- Shabala, S. Salinity and Programmed Cell Death: Unravelling Mechanisms for Ion Specific Signalling. J. Exp. Bot. 2009, 60, 709–712. [Google Scholar] [CrossRef]
- Kim, Y.; Wang, M.; Bai, Y.; Zeng, Z.; Guo, F.; Han, N.; Bian, H.; Wang, J.; Pan, J.; Zhu, M. Bcl-2 Suppresses Activation of VPEs by Inhibiting Cytosolic Ca2+ Level with Elevated K+ Efflux in NaCl-Induced PCD in Rice. Plant Physiol. Biochem. 2014, 80, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Minina, E.A.; Filonova, L.H.; Fukada, K.; Savenkov, E.I.; Gogvadze, V.; Clapham, D.; Sanchez-Vera, V.; Suarez, M.F.; Zhivotovsky, B.; Daniel, G.; et al. Autophagy and Metacaspase Determine the Mode of Cell Death in Plants. J. Cell Biol. 2013, 203, 917–927. [Google Scholar] [CrossRef]
- Bozhkov, P.V.; Suarez, M.F.; Filonova, L.H.; Daniel, G.; Zamyatnin, A.A.; Rodriguez-Nieto, S.; Zhivotovsky, B.; Smertenko, A. Cysteine Protease mcII-Pa Executes Programmed Cell Death during Plant Embryogenesis. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 14463–14468. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Yu, X.-H.; Wang, C.; Zhang, Q.; Liu, W.; McSweeney, S.; Shanklin, J.; Lam, E.; Liu, Q. Structural Basis for Ca2+-Dependent Activation of a Plant Metacaspase. Nat. Commun. 2020, 11, 2249. [Google Scholar] [CrossRef] [PubMed]
- Garcia, N.; Kalicharan, R.E.; Kinch, L.; Fernandez, J. Regulating Death and Disease: Exploring the Roles of Metacaspases in Plants and Fungi. Int. J. Mol. Sci. 2022, 24, 312. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.-Y.; Wang, Y.-J.; Jiao, J.-L.; Wang, W.-W.; Wang, H.-Z. The Metacaspase TaMCA-Id Negatively Regulates Salt-Induced Programmed Cell Death and Functionally Links With Autophagy in Wheat. Front. Plant Sci. 2022, 13, 904933. [Google Scholar] [CrossRef]
- Minina, E.A.; Smertenko, A.P.; Bozhkov, P.V. Vacuolar Cell Death in Plants: Metacaspase Releases the Brakes on Autophagy. Autophagy 2014, 10, 928–929. [Google Scholar] [CrossRef]
- Chen, H.; Dong, J.; Wang, T. Autophagy in Plant Abiotic Stress Management. Int. J. Mol. Sci. 2021, 22, 4075. [Google Scholar] [CrossRef]
- Koh, E.; Sunil, R.S.; Lam, H.Y.I.; Mutwil, M. Confronting the Data Deluge: How Artificial Intelligence Can Be Used in the Study of Plant Stress. Comput. Struct. Biotechnol. J. 2024, 23, 3454–3466. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep Learning in Agriculture: A Survey. Comput Electron Agric 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Khalifani, S.; Darvishzadeh, R.; Azad, N.; Seyed Rahmani, R. Prediction of Sunflower Grain Yield under Normal and Salinity Stress by RBF, MLP and, CNN Models. Ind Crops Prod 2022, 189, 115762. [Google Scholar] [CrossRef]
- Navarro, A.; Nicastro, N.; Costa, C.; Pentangelo, A.; Cardarelli, M.; Ortenzi, L.; Pallottino, F.; Cardi, T.; Pane, C. Sorting Biotic and Abiotic Stresses on Wild Rocket by Leaf-Image Hyperspectral Data Mining with an Artificial Intelligence Model. Plant Methods 2022, 18, 45. [Google Scholar] [CrossRef]
- Zhou, C.; Ye, H.; Sun, D.; Yue, J.; Yang, G.; Hu, J. An Automated, High-Performance Approach for Detecting and Characterizing Broccoli Based on UAV Remote-Sensing and Transformers: A Case Study from Haining, China. Int J Appl Earth Obs Geoinf 2022, 114, 103055. [Google Scholar] [CrossRef]
- Ho Tong Minh, D.; Ienco, D.; Gaetano, R.; Lalande, N.; Ndikumana, E.; Osman, F.; Maurel, P. Deep Recurrent Neural Networks for Winter Vegetation Quality Mapping via Multitemporal SAR Sentinel-1. IEEE Geosci. Remote Sens. Lett. 2018, 15, 464–468. [Google Scholar] [CrossRef]
- Atef, M.; Khattab, A.; Agamy, E.A.; Khairy, M.M. Deep Learning Based Time-Series Forecasting Framework for Olive Precision Farming. In Proceedings of the 2021 IEEE International Midwest Symposium on Circuits and Systems (MWSCAS); August 2021; pp. 1062–1065. [Google Scholar]
- Alhnaity, B.; Pearson, S.; Leontidis, G.; Kollias, S. Using Deep Learning to Predict Plant Growth and Yield in Greenhouse Environments. Acta Hortic. 2019, 1296, 55. [Google Scholar] [CrossRef]
- Zarbakhsh, S.; Shahsavar, A.R. Artificial Neural Network-Based Model to Predict the Effect of γ-Aminobutyric Acid on Salinity and Drought Responsive Morphological Traits in Pomegranate. Sci Rep 2022, 12, 16662. [Google Scholar] [CrossRef]
- Gao, G.; Tester, M.A.; Julkowska, M.M. The Use of High-Throughput Phenotyping for Assessment of Heat Stress-Induced Changes in Arabidopsis. Plant Phenomics 2020, 2020, 3723916. [Google Scholar] [CrossRef]
- Raza, A.; Gangurde, S.S.; Singh Sandhu, K.; Lv, Y. Omics-Assisted Crop Improvement under Abiotic Stress Conditions. Plant Stress 2024, 100626. [Google Scholar] [CrossRef]
- Meher, P.K.; Sahu, T.K.; Gupta, A.; Kumar, A.; Rustgi, S. ASRpro: A Machine-Learning Computational Model for Identifying Proteins Associated with Multiple Abiotic Stress in Plants. Plant Genome 2022, 17, e20259. [Google Scholar] [CrossRef]
- Pradhan, U.K.; Meher, P.K.; Naha, S.; Rao, A.R.; Kumar, U.; Pal, S.; Gupta, A. ASmiR: A Machine Learning Framework for Prediction of Abiotic Stress–Specific miRNAs in Plants. Funct Integr Genomics 2023, 23, 92. [Google Scholar] [CrossRef] [PubMed]
- Medina, C.A.; Hawkins, C.; Liu, X.-P.; Peel, M.; Yu, L.-X. Genome-Wide Association and Prediction of Traits Related to Salt Tolerance in Autotetraploid Alfalfa (Medicago sativa L.). Int J Mol Sci 2020, 21, 3361. [Google Scholar] [CrossRef] [PubMed]
- Medina, C.A.; Kaur, H.; Ray, I.; Yu, L.-X. Strategies to Increase Prediction Accuracy in Genomic Selection of Complex Traits in Alfalfa (Medicago sativa L.). Cells 2021, 10, 3372. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Ahn, H.; Lee, S.; Lee, C.-J.; Hur, J.; Jung, W.; Kim, S. StressGenePred: A Twin Prediction Model Architecture for Classifying the Stress Types of Samples and Discovering Stress-Related Genes in Arabidopsis. BMC Genomics 2019, 20, 949. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Chaudhary, P.; Taunk, J.; Singh, C.K.; Singh, D.; Tomar, R.S.S.; Aski, M.; Konjengbam, N.S.; Raje, R.S.; Singh, S.; et al. Fab Advances in Fabaceae for Abiotic Stress Resilience: From “Omics” to Artificial Intelligence. Int J Mol Sci 2021, 22, 10535. [Google Scholar] [CrossRef]
- Ullah, M.A.; Abdullah-Zawawi, M.-R.; Zainal-Abidin, R.-A.; Sukiran, N.L.; Uddin, M.I.; Zainal, Z. A Review of Integrative Omic Approaches for Understanding Rice Salt Response Mechanisms. Plants 2022, 11, 1430. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLOS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Tuncbag, N.; Gosline, S.J.C.; Kedaigle, A.; Soltis, A.R.; Gitter, A.; Fraenkel, E. Network-Based Interpretation of Diverse High-Throughput Datasets through the Omics Integrator Software Package. PLOS Comput. Biol. 2016, 12, e1004879. [Google Scholar] [CrossRef]
- Zoppi, J.; Guillaume, J.-F.; Neunlist, M.; Chaffron, S. MiBiOmics: An Interactive Web Application for Multi-Omics Data Exploration and Integration. BMC Bioinformatics 2021, 22, 6. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, B.; Li, G.; Chen, X.; Li, H.; Xu, X.; Chen, S.; He, W.; Xu, C.; Liu, L.; et al. An AI Agent for Fully Automated Multi-Omic Analyses. Adv. Sci. 2024, 2407094. [Google Scholar] [CrossRef]
- Wang, H.; Cimen, E.; Singh, N.; Buckler, E. Deep Learning for Plant Genomics and Crop Improvement. Curr. Opin. Plant Biol. 2020, 54, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Zaied, Y.B.; Zouabi, O. Impacts of Climate Change on Tunisian Olive Oil Output. Clim. Change 2016, 139, 535–549. [Google Scholar] [CrossRef]
- Harfouche, A.L.; Jacobson, D.A.; Kainer, D.; Romero, J.C.; Harfouche, A.H.; Scarascia Mugnozza, G.; Moshelion, M.; Tuskan, G.A.; Keurentjes, J.J.B.; Altman, A. Accelerating Climate Resilient Plant Breeding by Applying Next-Generation Artificial Intelligence. Trends Biotechnol. 2019, 37, 1217–1235. [Google Scholar] [CrossRef] [PubMed]
- Bey, N.E.; Maazoun, A.M.; Nahdi, O.; Krima, N.B.; Aounallah, M.K. Water Stress Indicators in Citrus, Olive and Apple Trees: A Review. J. Appl. Hortic. 2024, 26, 3–9. [Google Scholar] [CrossRef]
- Chartzoulakis, K.; Loupassaki, M.; Androulakis, I. Comparative Study on NaCl Salinity Tolerance of Six Olive Cultivars. Acta Hortic. 2002, 497–501. [Google Scholar] [CrossRef]
- Chartzoulakis, K.; Loupassaki, M.; Bertaki, M.; Androulakis, I. Effects of NaCl Salinity on Growth, Ion Content and CO2 Assimilation Rate of Six Olive Cultivars. Sci. Hortic. 2002, 96, 235–247. [Google Scholar] [CrossRef]
- Kchaou, H.; Larbi, A.; Gargouri, K.; Chaieb, M.; Morales, F.; Msallem, M. Assessment of Tolerance to NaCl Salinity of Five Olive Cultivars, Based on Growth Characteristics and Na+ and Cl− Exclusion Mechanisms. Sci. Hortic. 2010, 124, 306–315. [Google Scholar] [CrossRef]
- Soda, N.; Ephrath, J.E.; Dag, A.; Beiersdorf, I.; Presnov, E.; Yermiyahu, U.; Ben-Gal, A. Root Growth Dynamics of Olive (Olea europaea L.) Affected by Irrigation Induced Salinity. Plant Soil 2017, 411, 305–318. [Google Scholar] [CrossRef]
- Rugini, E.; Fedeli, E. Olive (Olea Europaea L.) as an Oilseed Crop. In Legumes and Oilseed Crops I.; Bajaj, Y.P.S., Ed.; Biotechnology in Agriculture and Forestry; Springer Berlin Heidelberg: Berlin, Heidelberg, 1990; ISBN 978-3-642-74450-1. [Google Scholar]
- Chartzoulakis, K.S. Salinity and Olive: Growth, Salt Tolerance, Photosynthesis and Yield. Agric. Water Manag. 2005, 78, 108–121. [Google Scholar] [CrossRef]
- Tattini, M.; Mergar, J.C.; Traversi, M.L. Responses of Olea Europaea to High Salinity : A Brief-Ecophysiological-Review. Adv. Hortic. Sci. 2008. [Google Scholar] [CrossRef]
- Regni, L.; Del Pino, A.M.; Mousavi, S.; Palmerini, C.A.; Baldoni, L.; Mariotti, R.; Mairech, H.; Gardi, T.; D’Amato, R.; Proietti, P. Behavior of Four Olive Cultivars During Salt Stress. Front. Plant Sci. 2019, 10, 867. [Google Scholar] [CrossRef]
- Loreto, F.; Centritto, M.; Chartzoulakis, K. Photosynthetic Limitations in Olive Cultivars with Different Sensitivity to Salt Stress: Photosynthetic Limitations and Salt Stress in Olive Cultivars. Plant Cell Environ. 2003, 26, 595–601. [Google Scholar] [CrossRef]
- Ben Abdallah, M.; Trupiano, D.; Polzella, A.; De Zio, E.; Sassi, M.; Scaloni, A.; Zarrouk, M.; Ben Youssef, N.; Scippa, G.S. Unraveling Physiological, Biochemical and Molecular Mechanisms Involved in Olive (Olea europaea L. Cv. Chétoui ) Tolerance to Drought and Salt Stresses. J. Plant Physiol. 2018, 220, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Sofo, A.; Dichio, B.; Xiloyannis, C.; Masia, A. Lipoxygenase Activity and Proline Accumulation in Leaves and Roots of Olive Trees in Response to Drought Stress. Physiol. Plant. 2004, 121, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Melgar, J.C.; Syvertsen, J.P.; Martínez, V.; García-Sánchez, F. The Relative Salt Tolerance of ‘Rangpur’ Seedlings and ‘Arbequina’ Olive Cuttings. Proc Fla State Hort Soc 2007, 120, 79–84. [Google Scholar]
- Moretti, S.; Francini, A.; Hernández, M.L.; Martínez-Rivas, J.M.; Sebastiani, L. Effect of Saline Irrigation on Physiological Traits, Fatty Acid Composition and Desaturase Genes Expression in Olive Fruit Mesocarp. Plant Physiol. Biochem. 2019, 141, 423–430. [Google Scholar] [CrossRef]
- Tattini, M.; Traversi, M. On the Mechanism of Salt Tolerance in Olive (Olea europaea L.) under Low- or High-Ca2+ Supply. Environ. Exp. Bot. 2009, 65, 72–81. [Google Scholar] [CrossRef]
- Cimato, A.; Castelli, S.; Tattini, M.; Traversi, M.L. An Ecophysiological Analysis of Salinity Tolerance in Olive. Environ. Exp. Bot. 2010, 68, 214–221. [Google Scholar] [CrossRef]
- Sodini, M.; Astolfi, S.; Francini, A.; Sebastiani, L. Multiple Linear Regression and Linear Mixed Models Identify Novel Traits of Salinity Tolerance in Olea europaea L. Tree Physiol. 2022, 42, 1029–1042. [Google Scholar] [CrossRef]
- Tadić, J.; Dumičić, G.; Veršić Bratinčević, M.; Vitko, S.; Radić Brkanac, S. Physiological and Biochemical Response of Wild Olive (Olea europaea subsp. europaea var. sylvestris) to Salinity. Front. Plant Sci. 2021, 12, 712005. [Google Scholar] [CrossRef]
- Goreta, S.; Bučević-Popović, V.; Pavela-Vrančić, M.; Perica, S. Salinity-induced Changes in Growth, Superoxide Dismutase Activity, and Ion Content of Two Olive Cultivars. J. Plant Nutr. Soil Sci. 2007, 170, 398–403. [Google Scholar] [CrossRef]
- Petridis, A.; Therios, I.; Samouris, G.; Tananaki, C. Salinity-Induced Changes in Phenolic Compounds in Leaves and Roots of Four Olive Cultivars (Olea europaea L.) and Their Relationship to Antioxidant Activity. Environ. Exp. Bot. 2012, 79, 37–43. [Google Scholar] [CrossRef]
- Rossi, L.; Borghi, M.; Francini, A.; Lin, X.; Xie, D.-Y.; Sebastiani, L. Salt Stress Induces Differential Regulation of the Phenylpropanoid Pathway in Olea europaea Cultivars Frantoio (Salt-Tolerant) and Leccino (Salt-Sensitive). J. Plant Physiol. 2016, 204, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Palm, E.R.; Salzano, A.M.; Vergine, M.; Negro, C.; Guidi Nissim, W.; Sabbatini, L.; Balestrini, R.; De Pinto, M.C.; Fortunato, S.; Gohari, G.; et al. Response to Salinity Stress in Four Olea europaea L. Genotypes: A Multidisciplinary Approach. Environ. Exp. Bot. 2024, 218, 105586. [Google Scholar] [CrossRef]
- Ahmed, Ch.B.; Rouina, B.B.; Sensoy, S.; Boukhris, M.; Abdallah, F.B. Changes in Gas Exchange, Proline Accumulation and Antioxidative Enzyme Activities in Three Olive Cultivars under Contrasting Water Availability Regimes. Environ. Exp. Bot. 2009, 67, 345–352. [Google Scholar] [CrossRef]
- Cordovilla, M. del P.; Aparicio, C.; Melendo, M.; Bueno, M. Exogenous Application of Indol-3-Acetic Acid and Salicylic Acid Improves Tolerance to Salt Stress in Olive Plantlets (Olea europaea L. cultivar Picual) in Growth Chamber Environments. Agronomy 2023, 13, 647. [Google Scholar] [CrossRef]
- Aparicio, C.; Urrestarazu, M.; Cordovilla, M.D.P. Comparative Physiological Analysis of Salinity Effects in Six Olive Genotypes. HortScience 2014, 49, 901–904. [Google Scholar] [CrossRef]
- Bashir, M.A.; Silvestri, C.; Coppa, E.; Brunori, E.; Cristofori, V.; Rugini, E.; Ahmad, T.; Hafiz, I.A.; Abbasi, N.A.; Nawaz Shah, M.K.; et al. Response of Olive Shoots to Salinity Stress Suggests the Involvement of Sulfur Metabolism. Plants 2021, 10, 350. [Google Scholar] [CrossRef]
- Mousavi, S.; Regni, L.; Bocchini, M.; Mariotti, R.; Cultrera, N.G.M.; Mancuso, S.; Googlani, J.; Chakerolhosseini, M.R.; Guerrero, C.; Albertini, E.; et al. Physiological, Epigenetic and Genetic Regulation in Some Olive Cultivars under Salt Stress. Sci. Rep. 2019, 9, 1093. [Google Scholar] [CrossRef]
- Tabatabaei, S.J. Salinity Stress and Olive: An Overview. Plant Stress 2007, 1, 105–112. [Google Scholar]
- Azimi, M.; Khoshzaman, T.; Taheri, M.; Dadras, A. Evaluation of Salinity Tolerance of Three Olive (Olea europaea L.) Cultivars. J. Cent. Eur. Agric. 2021, 22, 571–581. [Google Scholar] [CrossRef]
- Rahemi, M.; Karimi, S.; Sedaghat, S.; Ali Rostami, A. Physiological Responses of Olive Cultivars to Salinity Stress. Adv. Hortic. Sci. 2017, 31, 53–59. [Google Scholar] [CrossRef]
- Ayaz, M.; Varol, N.; Yolcu, S.; Pelvan, A.; Kaya, Ü.; Aydoğdu, E.; Bor, M.; Özdemir, F.; Türkan, İ. Three (Turkish) Olive Cultivars Display Contrasting Salt Stress-Coping Mechanisms under High Salinity. Trees 2021, 35, 1283–1298. [Google Scholar] [CrossRef]
- Chartzoulakis, K.S. The Potential of Saline and Residual Water Use in Olive Growing. Acta Hortic. 2014, 257–273. [Google Scholar] [CrossRef]
- Marin, L.; Benlloch, M.; Fernández-Escobar, R. Screening of Olive Cultivars for Salt Tolerance. Sci. Hortic. 1995, 64, 113–116. [Google Scholar] [CrossRef]
- Kchaou, H.; Larbi, A.; Gargouri, K.; Chaieb, M.; Morales, F.; Msallem, M. Assessment of Tolerance to NaCl Salinity of Five Olive Cultivars, Based on Growth Characteristics and Na+ and Cl− Exclusion Mechanisms. Sci. Hortic. 2010, 124, 306–315. [Google Scholar] [CrossRef]
- Aragüés, R.; Guillén, M.; Royo, A. Five-Year Growth and Yield Response of Two Young Olive Cultivars (Olea europaea L., Cvs. Arbequina and Empeltre) to Soil Salinity. Plant Soil 2010, 334, 423–432. [Google Scholar] [CrossRef]
- Bakshi, P. ; Amit; Wali, V.K. Mitigation Strategies of Abiotic Stress in Fruit Crops. In Abiotic & Biotic Stress Management in Plants, Ed.; CRC Press: London, 2022; ISBN 978-1-00-328198-6. [Google Scholar]
- Al-Absi, K.; Qrunfleh, M.; Abu-Sharar, T. Mechanism of Salt Tolerance of Two Olive Olea europaea L. Cultivars as Related to Electrolyte Concentration and Toxicity. Acta Hortic. 2003, 618, 281–290. [Google Scholar] [CrossRef]
- Perica, S.; Brkljača, M.; Goreta, S.; Romić, M. Vegetative Growth and Salt Accumulation of Six Olive Cultivars under Salt Stress. Acta Hortic. 2004, 664, 555–560. [Google Scholar] [CrossRef]
- AL-Ouda, A. Evaluation of the Response of Some Syrian Olive (Olea Europea L.) Cultivars to Salinity Stress.
- Bazakos, C.; Manioudaki, M.E.; Therios, I.; Voyiatzis, D.; Kafetzopoulos, D.; Awada, T.; Kalaitzis, P. Comparative Transcriptome Analysis of Two Olive Cultivars in Response to NaCl-Stress. PLoS ONE 2012, 7, e42931. [Google Scholar] [CrossRef]
- Bazakos, C.; Manioudaki, M.E.; Sarropoulou, E.; Spano, T.; Kalaitzis, P. 454 Pyrosequencing of Olive (Olea europaea L.) Transcriptome in Response to Salinity. PLOS ONE 2015, 10, e0143000. [Google Scholar] [CrossRef] [PubMed]
- Demir, S.; Cetinkaya, H. Effects of Saline Conditions on Polyphenol and Protein Content and Photosynthetic Response of Different Olive (Olea europaea L.). Appl. Ecol. Environ. Res. 2020, 18, 2599–2610. [Google Scholar] [CrossRef]
- Poury, N.; Seifi, E.; Alizadeh, M. Effects of Salinity and Proline on Growth and Physiological Characteristics of Three Olive Cultivars. Gesunde Pflanz. 2023, 75, 1169–1180. [Google Scholar] [CrossRef]
- El-Sayed Emtithal, H.; El-Said, M.E.; El-Sherif, A.H.; Sari El-Deen, S.A. Estudios químicos sobre la tolerancia a la sanidad de algunos cultivares. Olivae 1996, 64, 52–57. [Google Scholar]
- Mousavi, S.; Mariotti, R.; Valeri, M.C.; Regni, L.; Lilli, E.; Albertini, E.; Proietti, P.; Businelli, D.; Baldoni, L. Characterization of Differentially Expressed Genes under Salt Stress in Olive. Int. J. Mol. Sci. 2021, 23, 154. [Google Scholar] [CrossRef] [PubMed]
- Hassan, I.; Gaballah, M.; Bedour, A.; Proietti, P.; Regni, L. Salinity Stress Effects on Three Different Olive Cultivars and the Possibility of Their Cultivation in Reclaimed Lands. Plant Arch. 2020, 20, 2378–2382. [Google Scholar]
- Therios, I.N.; Misopolinos, N.D. Genotypic Response to Sodium Chloride Salinity of Four Major Olive Cultivars (Olea europea L.). Plant Soil 1988, 106, 105–111. [Google Scholar] [CrossRef]
- Skodra, C.; Michailidis, M.; Dasenaki, M.; Ganopoulos, I.; Thomaidis, N.S.; Tanou, G.; Molassiotis, A. Unraveling Salt-responsive Tissue-specific Metabolic Pathways in Olive Tree. Physiol. Plant. 2021, 173, 1643–1656. [Google Scholar] [CrossRef]
- Skodra, C.; Michailidis, M.; Moysiadis, T.; Stamatakis, G.; Ganopoulou, M.; Adamakis, I.-D.S.; Angelis, L.; Ganopoulos, I.; Tanou, G.; Samiotaki, M.; et al. Disclosing the Molecular Basis of Salinity Priming in Olive Trees Using Proteogenomic Model Discovery. Plant Physiol. 2023, 191, 1913–1933. [Google Scholar] [CrossRef]
- Briccoli-Bati, C.; Basta, P.; Tocci, C.; Turco, D. Influencia del riego con agua salobre en jóvenes plantas de oliva. Olivae 1994, 53, 35–38. [Google Scholar]
- Cruz, F.; Julca, I.; Gómez-Garrido, J.; Loska, D.; Marcet-Houben, M.; Cano, E.; Galán, B.; Frias, L.; Ribeca, P.; Derdak, S.; et al. Genome Sequence of the Olive Tree, Olea europaea. GigaScience 2016, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Julca, I.; Marcet-Houben, M.; Cruz, F.; Gómez-Garrido, J.; Gaut, B.S.; Díez, C.M.; Gut, I.G.; Alioto, T.S.; Vargas, P.; Gabaldón, T. Genomic Evidence for Recurrent Genetic Admixture during the Domestication of Mediterranean Olive Trees (Olea europaea L.). BMC Biol. 2020, 18, 148. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Ruiz, J.; Ramírez-Tejero, J.A.; Fernández-Pozo, N.; Leyva-Pérez, M. de la O.; Yan, H.; Rosa, R. de la; Belaj, A.; Montes, E.; Rodríguez-Ariza, M.O.; Navarro, F.; et al. Transposon Activation Is a Major Driver in the Genome Evolution of Cultivated Olive Trees (Olea europaea L.). Plant Genome 2020, 13, e20010. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.; Zhang, J.; Liu, X.; Lin, C.; Xin, H.; Xue, L.; Wang, C. De Novo Assembly of a New Olea europaea Genome Accession Using Nanopore Sequencing. Hortic. Res. 2021, 8, 64. [Google Scholar] [CrossRef]
- Lv, J.; Jiang, C.; Wu, W.; Mao, K.; Wei, Q.; Zheng, Y.; Gao, C.; Niu, Z.; Jin, G.; Zhang, R.; et al. The Gapless Genome Assembly and Multi-Omics Analyses Unveil a Pivotal Regulatory Mechanism of Oil Biosynthesis in the Olive Tree. Hortic. Res. 2024, 11, uhae168. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, J.; Peng, D.; Tian, Y.; Zhao, D.; Ni, W.; Long, J.; Li, J.; Zeng, Y.; Wu, Z.; et al. High-Quality Genome Assembly of Olea europaea subsp. cuspidata Provides Insights Into Its Resistance to Fungal Diseases in the Summer Rain Belt in East Asia. Front. Plant Sci. 2022, 13, 879822. [Google Scholar] [CrossRef]
- Wu, T.; Ma, T.; Xu, T.; Pan, L.; Zhang, Y.; Li, Y.; Ning, D. The De Novo Genome Assembly of Olea europaea subsp. cuspidata, a Widely Distributed Olive Close Relative. Front. Genet. 2022, 13, 868540. [Google Scholar] [CrossRef]
- Khadivi, A.; Mirheidari, F.; Saeidifar, A.; Moradi, Y. Morphological Characterizations of Olea europaea subsp. cuspidata. Genet. Resour. Crop Evol. 2024, 71, 1837–1853. [Google Scholar] [CrossRef]
- Yolcu, S.; Alavilli, H.; Ganesh, P.; Panigrahy, M.; Song, K. Salt and Drought Stress Responses in Cultivated Beets (Beta vulgaris L.) and Wild Beet (Beta maritima L.). Plants 2021, 10, 1843. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, D.; Liu, B.; Luo, W.; Lu, J.; Ma, T.; Wan, D. Transcriptome Dynamics of a Desert Poplar (Populus pruinosa) in Response to Continuous Salinity Stress. Plant Cell Rep. 2014, 33, 1565–1579. [Google Scholar] [CrossRef]
- Jamil, A.; Riaz, S.; Ashraf, M.; Foolad, M.R. Gene Expression Profiling of Plants under Salt Stress. Crit. Rev. Plant Sci. 2011, 30, 435–458. [Google Scholar] [CrossRef]
- Hoang, X.L.T.; Nhi, D.N.H.; Thu, N.B.A.; Thao, N.P.; Tran, L.-S.P. Transcription Factors and Their Roles in Signal Transduction in Plants under Abiotic Stresses. Curr. Genomics 2017, 18, 483–497. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) Transcription Factor Enhances Drought Resistance and Salt Tolerance in Rice. Proc. Natl. Acad. Sci. 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed]
- Cabello, J.V.; Lodeyro, A.F.; Zurbriggen, M.D. Novel Perspectives for the Engineering of Abiotic Stress Tolerance in Plants. Curr. Opin. Biotechnol. 2014, 26, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Rallo, L.; Barranco, D.; Díez, C.M.; Rallo, P.; Suárez, M.P.; Trapero, C.; Pliego-Alfaro, F. Strategies for Olive (Olea europaea L.) Breeding: Cultivated Genetic Resources and Crossbreeding. In Advances in Plant Breeding Strategies: Fruits; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer International Publishing: Cham, 2018; ISBN 978-3-319-91943-0. [Google Scholar]
- Jiang, X.; Zhang, W.; Fernie, A.R.; Wen, W. Combining Novel Technologies with Interdisciplinary Basic Research to Enhance Horticultural Crops. Plant J. 2022, 109, 35–46. [Google Scholar] [CrossRef]
- Sadder, M.T.; Ateyyeh, A.F.; Alswalmah, H.; Zakri, A.M.; Alsadon, A.A.; Al-Doss, A.A. Characterization of Putative Salinity-Responsive Biomarkers in Olive (Olea europaea L.). Plant Genet. Resour. Charact. Util. 2021, 19, 133–143. [Google Scholar] [CrossRef]
- Qin, Y.; Druzhinina, I.S.; Pan, X.; Yuan, Z. Microbially Mediated Plant Salt Tolerance and Microbiome-Based Solutions for Saline Agriculture. Biotechnol. Adv. 2016, 34, 1245–1259. [Google Scholar] [CrossRef]
- Fausto, C.; Mininni, A.N.; Sofo, A.; Crecchio, C.; Scagliola, M.; Dichio, B.; Xiloyannis, C. Olive Orchard Microbiome: Characterisation of Bacterial Communities in Soil-Plant Compartments and Their Comparison between Sustainable and Conventional Soil Management Systems. Plant Ecol. Divers. 2018, 11, 597–610. [Google Scholar] [CrossRef]
- Costa, D.; Fernandes, T.; Martins, F.; Pereira, J.A.; Tavares, R.M.; Santos, P.M.; Baptista, P.; Lino-Neto, T. Illuminating Olea europaea L. Endophyte Fungal Community. Microbiol. Res. 2021, 245, 126693. [Google Scholar] [CrossRef]
- Liu, H.; Brettell, L.E.; Qiu, Z.; Singh, B.K. Microbiome-Mediated Stress Resistance in Plants. Trends Plant Sci. 2020, 25, 733–743. [Google Scholar] [CrossRef]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2020, 10, 1741. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Dias, M.C.; Freitas, H. Drought and Salinity Stress Responses and Microbe-Induced Tolerance in Plants. Front. Plant Sci. 2020, 11, 591911. [Google Scholar] [CrossRef] [PubMed]
- Chialva, M.; De Rose, S.; Novero, M.; Lanfranco, L.; Bonfante, P. Plant Genotype and Seasonality Drive Fine Changes in Olive Root Microbiota. Curr. Plant Biol. 2021, 28, 100219. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-Microbiome Interactions: From Community Assembly to Plant Health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Mina, D.; Pereira, J.A.; Lino-Neto, T.; Baptista, P. Epiphytic and Endophytic Bacteria on Olive Tree Phyllosphere: Exploring Tissue and Cultivar Effect. Microb. Ecol. 2020, 80, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Fernández-González, A.J.; Villadas, P.J.; Gómez-Lama Cabanás, C.; Valverde-Corredor, A.; Belaj, A.; Mercado-Blanco, J.; Fernández-López, M. Defining the Root Endosphere and Rhizosphere Microbiomes from the World Olive Germplasm Collection. Sci. Rep. 2019, 9, 20423. [Google Scholar] [CrossRef]
- Malacrinò, A.; Mosca, S.; Li Destri Nicosia, M.G.; Agosteo, G.E.; Schena, L. Plant Genotype Shapes the Bacterial Microbiome of Fruits, Leaves, and Soil in Olive Plants. Plants 2022, 11, 613. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Li Destri Nicosia, M.G.; Cacciola, S.O.; Droby, S.; Schena, L. Metabarcoding Analysis of Fungal Diversity in the Phyllosphere and Carposphere of Olive (Olea europaea). PLOS ONE 2015, 10, e0131069. [Google Scholar] [CrossRef]
- Caliz, J.; Montes-Borrego, M.; Triadó-Margarit, X.; Metsis, M.; Landa, B.B.; Casamayor, E.O. Influence of Edaphic, Climatic, and Agronomic Factors on the Composition and Abundance of Nitrifying Microorganisms in the Rhizosphere of Commercial Olive Crops. PLOS ONE 2015, 10, e0125787. [Google Scholar] [CrossRef]
- Sofo, A.; Ricciuti, P.; Fausto, C.; Mininni, A.N.; Crecchio, C.; Scagliola, M.; Malerba, A.D.; Xiloyannis, C.; Dichio, B. The Metabolic and Genetic Diversity of Soil Bacterial Communities Depends on the Soil Management System and C/N Dynamics: The Case of Sustainable and Conventional Olive Groves. Appl. Soil Ecol. 2019, 137, 21–28. [Google Scholar] [CrossRef]
- Müller, H.; Berg, C.; Landa, B.B.; Auerbach, A.; Moissl-Eichinger, C.; Berg, G. Plant Genotype-Specific Archaeal and Bacterial Endophytes but Similar Bacillus Antagonists Colonize Mediterranean Olive Trees. Front. Microbiol. 2015, 6, 138. [Google Scholar] [CrossRef] [PubMed]
- Vita, F.; Sabbatini, L.; Sillo, F.; Ghignone, S.; Vergine, M.; Guidi Nissim, W.; Fortunato, S.; Salzano, A.M.; Scaloni, A.; Luvisi, A.; et al. Salt Stress in Olive Tree Shapes Resident Endophytic Microbiota. Front. Plant Sci. 2022, 13, 992395. [Google Scholar] [CrossRef] [PubMed]
- Fernández-González, A.J.; Ramírez-Tejero, J.A.; Nevado-Berzosa, M.P.; Luque, F.; Fernández-López, M.; Mercado-Blanco, J. Coupling the Endophytic Microbiome with the Host Transcriptome in Olive Roots. Comput. Struct. Biotechnol. J. 2021, 19, 4777–4789. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Meng, Y.; Zhao, M.; Wang, M.; Wang, C.; Dong, J.; Fan, W.; Xu, F.; Wang, D.; Xie, Z. Advances and Mechanisms of Fungal Symbionts in Improving the Salt Tolerance of Crops. Plant Sci. 2024, 349, 112261. [Google Scholar] [CrossRef]
- Martins, F.; Pereira, J.A.; Bota, P.; Bento, A.; Baptista, P. Fungal Endophyte Communities in Above- and Belowground Olive Tree Organs and the Effect of Season and Geographic Location on Their Structures. Fungal Ecol. 2016, 20, 193–201. [Google Scholar] [CrossRef]
- Fernández-González, A.J.; Wentzien, N.M.; Villadas, P.J.; Valverde-Corredor, A.; Lasa, A.V.; Gómez-Lama Cabanás, C.; Mercado-Blanco, J.; Fernández-López, M. Comparative Study of Neighboring Holm Oak and Olive Trees-Belowground Microbial Communities Subjected to Different Soil Management. PLOS ONE 2020, 15, e0236796. [Google Scholar] [CrossRef]
- Llimós, M.; Segarra, G.; Sancho-Adamson, M.; Trillas, M.I.; Romanyà, J. Impact of Olive Saplings and Organic Amendments on Soil Microbial Communities and Effects of Mineral Fertilization. Front. Microbiol. 2021, 12, 653027. [Google Scholar] [CrossRef]
- Sofo, A.; Palese, A.M.; Casacchia, T.; Xiloyannis, C. Sustainable Soil Management in Olive Orchards. In Emerging Technologies and Management of Crop Stress Tolerance; Elsevier, 2014; pp. 471–483 ISBN 978-0-12-800875-1.
- Kumar, V.; Kumar, P.; Khan, A. Optimization of PGPR and Silicon Fertilization Using Response Surface Methodology for Enhanced Growth, Yield and Biochemical Parameters of French Bean (Phaseolus vulgaris L.) under Saline Stress. Biocatal. Agric. Biotechnol. 2020, 23, 101463. [Google Scholar] [CrossRef]
- Roy, S.; Chakraborty, A.P.; Chakraborty, R. Understanding the Potential of Root Microbiome Influencing Salt-tolerance in Plants and Mechanisms Involved at the Transcriptional and Translational Level. Physiol. Plant. 2021, 173, 1657–1681. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Abrantes, I.; Barra Caracciolo, A.; Bevivino, A.; Ciancio, A.; Grenni, P.; Hrynkiewicz, K.; Kredics, L.; Proença, D.N. Belowground Microbiota and the Health of Tree Crops. Front. Microbiol. 2018, 9, 1006. [Google Scholar] [CrossRef]
- Costa-Gutierrez, S.B.; Lami, M.J.; Santo, M.C.C.-D.; Zenoff, A.M.; Vincent, P.A.; Molina-Henares, M.A.; Espinosa-Urgel, M.; de Cristóbal, R.E. Plant Growth Promotion by Pseudomonas putida KT2440 under Saline Stress: Role of eptA. Appl. Microbiol. Biotechnol. 2020, 104, 4577–4592. [Google Scholar] [CrossRef] [PubMed]
- Vives-Peris, V.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Salt Stress Alleviation in Citrus Plants by Plant Growth-Promoting Rhizobacteria Pseudomonas putida and Novosphingobium Sp. Plant Cell Rep. 2018, 37, 1557–1569. [Google Scholar] [CrossRef] [PubMed]
- Raupp, P.; Carrillo, Y.; Nielsen, U.N. Soil Health to Enhance Ecological Restoration and Conservation. J. Sustain. Agric. Environ. 2024, 3, e70022. [Google Scholar] [CrossRef]
- Kapoor, A.; Sharma, R.; Kumar, A.; Sepehya, S. Biochar as a Means to Improve Soil Fertility and Crop Productivity: A Review. J. Plant Nutr. 2022, 45, 2380–2388. [Google Scholar] [CrossRef]
- Khan, Z.; Jan, R.; Asif, S.; Farooq, M.; Jang, Y.-H.; Kim, E.-G.; Kim, N.; Kim, K.-M. Exogenous Melatonin Induces Salt and Drought Stress Tolerance in Rice by Promoting Plant Growth and Defense System. Sci. Rep. 2024, 14, 1214. [Google Scholar] [CrossRef]
- Ding, Z.; Kheir, A.M.S.; Ali, M.G.M.; Ali, O.A.M.; Abdelaal, A.I.N.; Lin, X.; Zhou, Z.; Wang, B.; Liu, B.; He, Z. The Integrated Effect of Salinity, Organic Amendments, Phosphorus Fertilizers, and Deficit Irrigation on Soil Properties, Phosphorus Fractionation and Wheat Productivity. Sci. Rep. 2020, 10, 2736. [Google Scholar] [CrossRef]
- Khan, I.; Mahmood, S.; Chattha, M.U.; Bilal Chattha, M.; Ahmad, S.; Awan, M.I.; Alqahtani, F.M.; Hashem, M.; Alhaithloul, H.A.S.; Qari, S.H.; et al. Organic Amendments Improved the Productivity and Bio-Fortification of Fine Rice by Improving Physiological Responses and Nutrient Homeostasis under Salinity Stress. Plants 2023, 12, 1644. [Google Scholar] [CrossRef]
- Chartzoulakis, K.; Psarras, G.; Vemmos, S.; Loupassaki, M.; Bertaki, M. Response of Two Olive Cultivars to Salt Stress and Potassium Supplement. J. Plant Nutr. 2006, 29, 2063–2078. [Google Scholar] [CrossRef]
- Ziskin, R.; Dag, A.; Yermiyahu, U.; Levy, G.J. Different Amendments for Combating Soil Sodicity in an Olive Orchard. Agric. Water Manag. 2024, 299, 108837. [Google Scholar] [CrossRef]
- Chehab, H.; Tekaya, M.; Hajlaoui, H.; Abdelhamid, S.; Gouiaa, M.; Sfina, H.; Chihaoui, B.; Boujnah, D.; Mechri, B. Complementary Irrigation with Saline Water and Soil Organic Amendments Modified Soil Salinity, Leaf Na+, Productivity and Oil Phenols of Olive Trees (Cv. Chemlali) Grown under Semiarid Conditions. Agric. Water Manag. 2020, 237, 106183. [Google Scholar] [CrossRef]
- Naeini, N.R.; Esna-Ashari, M.; Khoshgoftarmanesh, A.; Cheraghi, H.; Hatami, A.; Mirzapour, M. The Effect of Zinc Nutrition on Two Olive (Olea europaea L.) Cultivars Components and Alleviate Oxidative Damage in Salinity Conditions. J. Med. Plants Byprod. 2016, 1, 23–31. [Google Scholar]
- Rodriguez, R.J.; Henson, J.; Van Volkenburgh, E.; Hoy, M.; Wright, L.; Beckwith, F.; Kim, Y.-O.; Redman, R.S. Stress Tolerance in Plants via Habitat-Adapted Symbiosis. ISME J. 2008, 2, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Kumawat, K.C.; Razdan, N.; Saharan, K. Rhizospheric Microbiome: Bio-Based Emerging Strategies for Sustainable Agriculture Development and Future Perspectives. Microbiol. Res. 2022, 254, 126901. [Google Scholar] [CrossRef] [PubMed]
- Kumawat, K.C.; Nagpal, S.; Sharma, P. Potential of Plant Growth-Promoting Rhizobacteria-Plant Interactions in Mitigating Salt Stress for Sustainable Agriculture: A Review. Pedosphere 2022, 32, 223–245. [Google Scholar] [CrossRef]
- Barnawal, D.; Bharti, N.; Pandey, S.S.; Pandey, A.; Chanotiya, C.S.; Kalra, A. Plant Growth-Promoting Rhizobacteria Enhance Wheat Salt and Drought Stress Tolerance by Altering Endogenous Phytohormone Levels and TaCTR1/TaDREB2 Expression. Physiol. Plant. 2017, 161, 502–514. [Google Scholar] [CrossRef]
- Porras-Soriano, A.; Soriano-Martín, M.L.; Porras-Piedra, A.; Azcón, R. Arbuscular Mycorrhizal Fungi Increased Growth, Nutrient Uptake and Tolerance to Salinity in Olive Trees under Nursery Conditions. J. Plant Physiol. 2009, 166, 1350–1359. [Google Scholar] [CrossRef]
- Galla, G.; Barcaccia, G.; Ramina, A.; Collani, S.; Alagna, F.; Baldoni, L.; Cultrera, N.G.; Martinelli, F.; Sebastiani, L.; Tonutti, P. Computational Annotation of Genes Differentially Expressed along Olive Fruit Development. BMC Plant Biol. 2009, 9, 128. [Google Scholar] [CrossRef]
- Munoz-Merida, A.; Gonzalez-Plaza, J.J.; Canada, A.; Blanco, A.M.; Garcia-Lopez, M. d. C.; Rodriguez, J.M.; Pedrola, L.; Sicardo, M.D.; Hernandez, M.L.; De la Rosa, R.; et al. De Novo Assembly and Functional Annotation of the Olive (Olea europaea) Transcriptome. DNA Res. 2013, 20, 93–108. [Google Scholar] [CrossRef]
- Asins, M.J.; Bullones, A.; Raga, V.; Romero-Aranda, M.R.; Espinosa, J.; Triviño, J.C.; Bernet, G.P.; Traverso, J.A.; Carbonell, E.A.; Claros, M.G.; et al. Combining Genetic and Transcriptomic Approaches to Identify Transporter-Coding Genes as Likely Responsible for a Repeatable Salt Tolerance QTL in Citrus. Int. J. Mol. Sci. 2023, 24, 15759. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Zheng, Y.; Snyder, S.I.; Nicolas, P.; Shinozaki, Y.; Fei, Z.; Catala, C.; Giovannoni, J.J.; Rose, J.K.C.; Mueller, L.A. The Tomato Expression Atlas. Bioinforma. Oxf. Engl. 2017, 33, 2397–2398. [Google Scholar] [CrossRef]
- Bag, P.; Lihavainen, J.; Delhomme, N.; Riquelme, T.; Robinson, K.M.; Jansson, S. An Atlas of the Norway Spruce Needle Seasonal Transcriptome. Plant J. 2021, 108, 1815–1829. [Google Scholar] [CrossRef] [PubMed]
- Toufighi, K.; Brady, S.M.; Austin, R.; Ly, E.; Provart, N.J. The Botany Array Resource: E-Northerns, Expression Angling, and Promoter Analyses: The Botany Array Resource. Plant J. 2005, 43, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Pozo, N.; Haas, F.B.; Meyberg, R.; Ullrich, K.K.; Hiss, M.; Perroud, P.-F.; Hanke, S.; Kratz, V.; Powell, A.F.; Vesty, E.F.; et al. PEATmoss (Physcomitrella Expression Atlas Tool): A Unified Gene Expression Atlas for the Model Plant Physcomitrella patens. Plant J. Cell Mol. Biol. 2020, 102, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Ollé, A.; Bullones, A.; Hormaza, J.I.; Mueller, L.A.; Fernandez-Pozo, N. MangoBase: A Genomics Portal and Gene Expression Atlas for Mangifera indica. Plants 2023, 12, 1273. [Google Scholar] [CrossRef]
- Bullones, A.; Castro, A.J.; Lima-Cabello, E.; Alché, J.D.D.; Luque, F.; Claros, M.G.; Fernandez-Pozo, N. OliveAtlas: A Gene Expression Atlas Tool for Olea europaea. Plants 2023, 12, 1274. [Google Scholar] [CrossRef]
- Cruz, A.C.; Luvisi, A.; De Bellis, L.; Ampatzidis, Y. X-FIDO: An Effective Application for Detecting Olive Quick Decline Syndrome with Deep Learning and Data Fusion. Front. Plant Sci. 2017, 8, 1741. [Google Scholar] [CrossRef]
- Di Nisio, A.; Adamo, F.; Acciani, G.; Attivissimo, F. Fast Detection of Olive Trees Affected by Xylella fastidiosa from UAVs Using Multispectral Imaging. Sensors 2020, 20. [Google Scholar] [CrossRef]
- Sesli, M.; Yegenoğlu, E.D.; Altıntaa, V. Determination of Olive Cultivars by Deep Learning and ISSR Markers. J Env. Biol 2020, 41, 426–431. [Google Scholar] [CrossRef]
| Cultivar | Reference | Country | Salt treatment | Treatment time |
|---|---|---|---|---|
| Salt-tolerant cultivars | ||||
| Abou-Satl | [161] | Syria | 12 dS/m NaCl | 90 days |
| Amigdalilolia | [162] | Iran | 12 dS/m NaCl | 90 days |
| Ayvalık | [163] | Turkey | 300 mM NaCl | 30 days |
| Barnea | [164] | Israel | 7.5 dS/m EC | 9 years |
| Cañivano | [165] | Spain | 100 mM NaCl | 49 days |
| Chemlali | [164] [166] |
Tunisia | brackish water | 8 months |
| 200 mM NaCl | 5 months | |||
| Dakal | [162] | Iran | 12 dS/m NaCl | 90 days |
| Empeltre | [167] | Spain | 10 dS/m Eciw | 4 years |
| Escarabajuelo | [165] | Spain | 100 mM NaCl | 49 days |
| Frantoio | [168] | Italy | 200 mM NaCl | 60 days |
| [169] | 100 molc/m3 SIW | ND | ||
| [157] | 200 mM NaCl | 84 days | ||
| [154] | 200 mM NaCl | 56 days | ||
| Hamed | [164] | Egypt | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years |
| Istarska bjelica | [170] | Croatia | 100 mM NaCl | 70 days |
| Jabaluna | [165] | Spain | 100 mM NaCl | 49 days |
| Jlot | [171] | Syria | 8000 ppm NaCl | ND |
| Kalamata | [164] | Greece | 200 mM NaCl | 5 months |
| Kalamon | [172] | Greece | 120 mM NaCl | 90 days |
| [173] | 120 mM NaCl | 90 days | ||
| Kerkiras | [164] | Greece | ND | ND |
| Kilis | [163] | Turkey | 300 mM NaCl | 30 days |
| Kilis Yağlık | [174] | Turkey | 150 mM NaCl | 4 months |
| Koroneiki I38 | [175] | Spain | 200 mM NaCl | 5 months |
| Kothreiki | [164] | Greece | 200 mM NaCl | 5 months |
| Lechín de Sevilla | [165] | Spain | 100 mM NaCl | 49 days |
| Lianolia | [164] | Greece | ND | ND |
| Megaritiki | [168] | Greece | 150 meq NaCl | 4 months |
| [135] | 200 mM NaCl | 5 months | ||
| Nevadillo | [165] | Spain | 100 mM NaCl | 49 days |
| Oblica | [170] | Croatia | 100 mM NaCl | 70 days |
| Ocal | [157] | Spain | 200 mM NaCl | 84 days |
| Pocama | [176] | Egypt | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years |
| Royal de Cazorla | [141] | Spain | 100 mM and 200 mM NaCl | 8 months |
| [159] | 100 mM and 200 mM NaCl | 60 days | ||
| [177] | 200 mM NaCl | 8 months | ||
| Verdale | [176] | France | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years |
| Intermediate tolerance to salt | ||||
| Adramitini | [164] | Greece | ND | ND |
| Aggezi | [164] | Egypt | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years |
| Alameño | [165] | Spain | 100 mM NaCl | 49 days |
| Arbosana I43 | [166] | Spain | 200 mM NaCl | 5 months |
| Blanqueta | [165] | Spain | 100 mM NaCl | 49 days |
| Cañivano Negro | [165] | Spain | 100 mM NaCl | 49 days |
| Carolea | [168] | Italy | 14.97 mS/cm | 11 months |
| Casta Cabra | [157] | Spain | 200 mM NaCl | 84 days |
| Changlot Real | [165] | Spain | 100 mM NaCl | 49 days |
| Chorruo | [165] | Spain | 100 mM NaCl | 49 days |
| Coratina | [168] | Italy | 200 mM NaCl | 60 days |
| Cornicabra | [157] | Spain | 200 mM NaCl | 84 days |
| Dezful | [162] | Iran | 12 dS/m NaCl | 90 days |
| Gordal Sevillana | [165] | Spain | 100 mM NaCl | 49 days |
| Hojiblanca | [165] | Spain | 100 mM NaCl | 49 days |
| Khaisi | [171] | Syria | 8000 ppm NaCl | ND |
| Lechín de Granada | [165] | Spain | 100 mM NaCl | 49 days |
| Manzanilla de Sevilla | [165] | Spain | 100 mM NaCl | 49 days |
| Manzanillo | [168] | Spain | 60 mM NaCl | 115 days |
| [178] | 4000 mg/L MgSO4, CaSO4, NaCl, MgCl2 and CaCO3 | 9 months | ||
| [176] | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years | ||
| Maraiolo | [164] | Italy | ND | ND |
| Maurino | [164] | Italy | 200 mM NaCl | 60 days |
| Moraiolo | [168] | Italy | 200 mM NaCl | 60 days |
| Mostazal | [164] | Peru | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years |
| Nabali Muhassan | [164] | Jordan | 100 molc/m3 SIW | ND |
| Nizip Yağlik | [174] | Turkey | 150 mM NaCl | 4 months |
| Oblonga | [165] | Italy | 100 mM NaCl | 49 days |
| Redondil | [165] | Spain | 100 mM NaCl | 49 days |
| Toffahi | [164] | Egypt | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years |
| Tokhm-e-Kabki | [162] | Iran | 12 dS/m NaCl | 90 days |
| Valanolia | [164] | Greece | ND | ND |
| Verdial de Vélez-Málaga | [165] | Spain | 100 mM NaCl | 49 days |
| Wardan | [164] | Egypt | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years |
| Zorzariega | [165] | Spain | 100 mM NaCl | 49 days |
| Salt-sensitive cultivars | ||||
| Aggezi Shami | [178] | Egypt | 4000 mg/L MgSO4, CaSO4, NaCl, MgCl2 and CaCO3 | 275 days |
| Aguromanaki | [164] | Greece | ND | ND |
| Arbequina I18 | [166] | Spain | 200 mM NaCl | 5 months |
| Arbosana | [175] | Spain | 200 mM NaCl | 5 months |
| Bezeri | [171] | Syria | 8000 ppm NaCl | ND |
| Bouteillan | [164] | France | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years |
| Buža | [170] | Croatia | 100 mM NaCl | 70 days |
| Chondrolia Chalkidikis | [172] | Greece | 120 mM NaCl | 90 days |
| [179] | 150 meq NaCl | 4 months | ||
| [172] | 120 mM NaCl | 90 days | ||
| [180] | 75 mM NaCl | 45 days | ||
| [181] | 75 mM and 150 mM NaCl | 45 days | ||
| Cobrançosa | [165] | Portugal | 100 mM NaCl | 49 days |
| Conservalia | [162] | Iran | 12 dS/m NaCl | 90 days |
| Drobnica | [170] | Croatia | 100 mM NaCl | 70 days |
| Fadaq86 | [177] | Iran | 200 mM NaCl | 8 months |
| Galego | [165] | Spain | 100 mM NaCl | 49 days |
| Khederi | [171] | Syria | 8000 ppm NaCl | ND |
| Lastovka | [170] | Croatia | 100 mM NaCl | 70 days |
| Leccino | [168] | Italy | 200 mM NaCl | 60 days |
| [170] | 100 mM NaCl | 70 days | ||
| [154] | 200 mM NaCl | 56 days | ||
| Mastoidis | [134] | Greece | 200 mM NaCl | 5 months |
| Meski | [165] | Tunisia | 100 mM NaCl | 49 days |
| Mission | [176] | US | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years |
| Nabal | [164] | Egypt | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years |
| Nocellara del Belice | [182] | Italy | 14.97 mS/cm | 11 months |
| Pajarero | [168] | Spain | 100 mM NaCl | 49 days |
| Rosciola | [176] | Italy | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years |
| Shiraz | [162] | Iran | 12 dS/m NaCl | 90 days |
| Sourani | [171] | Syria | 8000 ppm NaCl | ND |
| Throubolia | [164] | Greece | ND | ND |
| Cultivar | Tolerant | Intermediate | Sensitive | Country | Salt treatment | Treatment time |
|---|---|---|---|---|---|---|
| Amphissis | [179] | Greece | 150 meq NaCl | 4 months | ||
| [134] | 200 mM NaCl | 5 months | ||||
| Arbequina | [168] | Spain | 100 mM NaCl | 49 days | ||
| [167] | 10 dS/m Eciw | 4 years | ||||
| [175] | 200 mM NaCl | 5 months | ||||
| [159] | 100 mM and 200 mM NaCl | 60 days | ||||
| [161] | 12 dS/m NaCl | 90 days | ||||
| Chétoui | [166] | Tunisia | 200 mM NaCl | 5 months | ||
| [165] | 100 mM NaCl | 49 days | ||||
| Gemlik | [163] | Turkey | 300 mM NaCl | 30 days | ||
| [174] | 150 mM NaCl | 4 months | ||||
| Koroneiki | [176] | Greece | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years | ||
| [179] | 150 meq NaCl | 4 months | ||||
| [159] | 100 mM and 200 mM NaCl | 60 days | ||||
| [141] | 100 mM and 200 mM NaCl | 8 months | ||||
| [177] | 200 mM NaCl | 8 months | ||||
| [166] | 200 mM NaCl | 5 months | ||||
| [134] | 200 mM NaCl | 5 months | ||||
| Picual | [168] | Spain | 100 mM NaCl | 49 days | ||
| [178] | 4000 mg/L MgSO4, CaSO4, NaCl, MgCl2 and CaCO3 | 9 months | ||||
| [176] | 6000 ppm NaCl, NaSO4, CaCl2 and MgSO4 | 2 years | ||||
| [159] | 100 mM and 200 mM NaCl | 60 days | ||||
| [177] | 200 mM NaCl | 8 months | ||||
| [157] | 200 mM NaCl | 84 days | ||||
| Picudo | [157] | Spain | 200 mM NaCl | 84 days | ||
| [165] | 100 mM NaCl | 49 days | ||||
| Zard | [161] | Iran | 12 dS/m NaCl | 90 days | ||
| [162] | 12 dS/m NaCl | 90 days |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





