Submitted:
28 November 2024
Posted:
29 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
- Sampling and characterization of buttermilk;
- Batch experiments of protein hydrolysis using different enzymes to select the best enzyme for the operation of the MBR;
- Protein hydrolysis and separation of peptides in the MBR.
2.1. Sampling and Characterization of Buttermilk
2.2. Batch Experiments of Protein Hydrolysis Using Different Enzymes
2.3. Enzymatic Hydrolysis and Separation of Peptides in the Membrane Bioreactor
2.3.1. MBR Operation
2.3.2. Peptide Characterization
2.3.3. Data Analysis
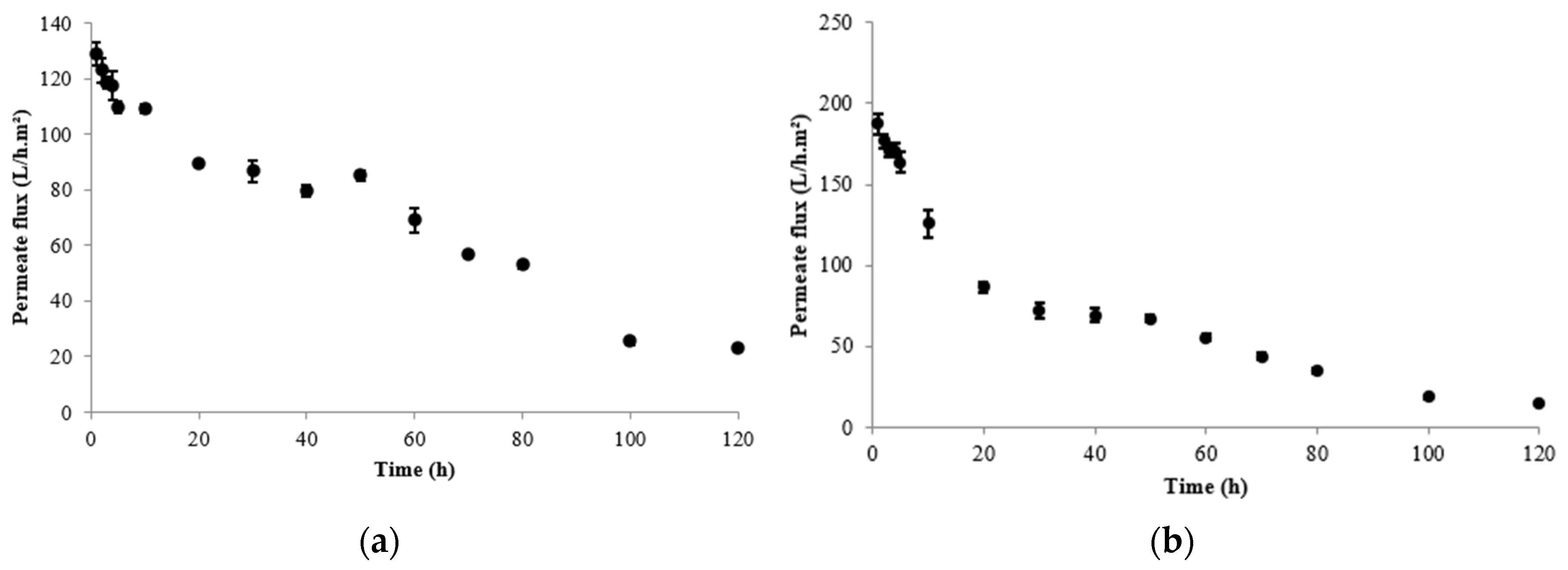
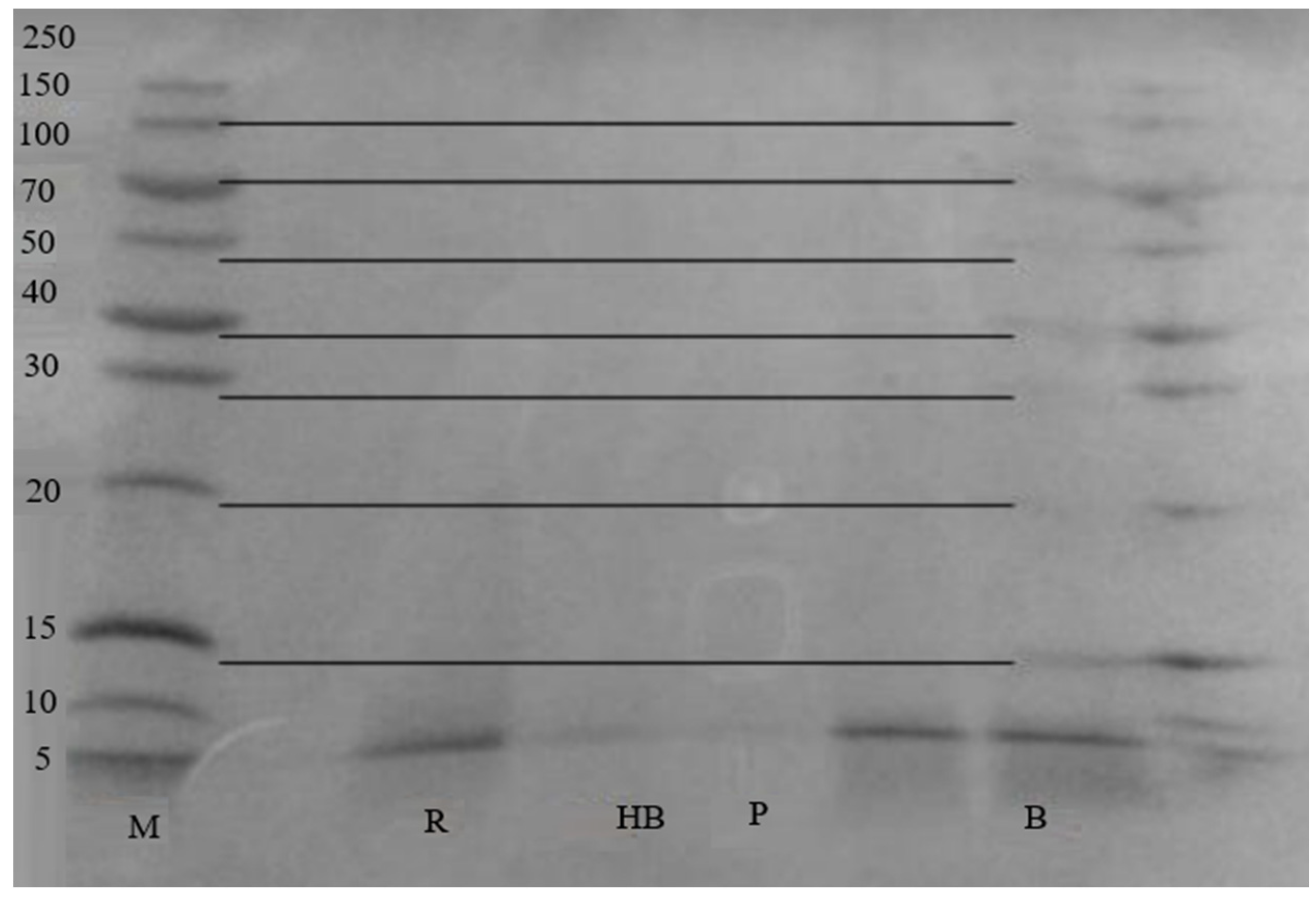
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pereira, C.; Gomes, D.; Dias, S.; Santos, S.; Pires, A.; Viegas, J. Impact of Probiotic and Bioprotective Cultures on the Quality and Shelf Life of Butter and Buttermilk. Dairy 2024, Vol. 5, Pages 625-643 2024, 5, 625–643. [Google Scholar] [CrossRef]
- Astaire, J.C.; Ward, R.; German, J.B.; Jiménez-Flores, R. Concentration of Polar MFGM Lipids from Buttermilk by Microfiltration and Supercritical Fluid Extraction. J Dairy Sci 2003, 86, 2297–2307. [Google Scholar] [CrossRef] [PubMed]
- Sakkas, L.; Evageliou, V.; Igoumenidis, P.; Moatsou, G. Properties of Sweet Buttermilk Released from the Churning of Cream Separated from Sheep or Cow Milk or Sheep Cheese Whey: Effect of Heat Treatment and Storage of Cream. Foods 2022, Vol. 11, Page 465 2022, 11, 465. [Google Scholar] [CrossRef] [PubMed]
- Golkar, A.; Milani, J.M.; Vasiljevic, T. Altering Allergenicity of Cow’s Milk by Food Processing for Applications in Infant Formula. Crit Rev Food Sci Nutr 2019, 59, 159–172. [Google Scholar] [CrossRef]
- Xu, D.; Yuan, F.; Jiang, J.; Wang, X.; Hou, Z.; Gao, Y. Structural and Conformational Modification of Whey Proteins Induced by Supercritical Carbon Dioxide. Innovative Food Science and Emerging Technologies 2011, 12, 32–37. [Google Scholar] [CrossRef]
- Cheung, I.W.Y.; Cheung, L.K.Y.; Tan, N.Y.; Li-Chan, E.C.Y. The Role of Molecular Size in Antioxidant Activity of Peptide Fractions from Pacific Hake (Merluccius Productus) Hydrolysates. Food Chem 2012, 134, 1297–1306. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, C.; Ren, Y.; Wang, C.; Tian, F. What Are the Ideal Properties for Functional Food Peptides with Antihypertensive Effect? A Computational Peptidology Approach. Food Chem 2013, 141, 2967–2973. [Google Scholar] [CrossRef]
- Marson, G.V.; Belleville, M.P.; Lacour, S.; Hubinger, M.D. Membrane Fractionation of Protein Hydrolysates from By-Products: Recovery of Valuable Compounds from Spent Yeasts. Membranes 2021, Vol. 11, Page 23 2020, 11, 23. [Google Scholar] [CrossRef]
- Faucher, M.; Geoffroy, T.R.; Thibodeau, J.; Gaaloul, S.; Bazinet, L. Semi-Industrial Production of a DPP-IV and ACE Inhibitory Peptide Fraction from Whey Protein Concentrate Hydrolysate by Electrodialysis with Ultrafiltration Membrane. Membranes 2022, Vol. 12, Page 409 2022, 12, 409. [Google Scholar] [CrossRef]
- Sbeghen, A.L.; Lira, A.L.; Fernandes, I.A.; Steffens, C.; Brião, V.B.; Zeni, J.; Steffens, J. Use of Ultrafiltration in the Separation of Hydrolysates from Mechanically Separated Chicken Meat and Evaluation of Antioxidant Activity. J Food Process Eng 2022, 45, e14151. [Google Scholar] [CrossRef]
- Nath, A.; Csighy, A.; Eren, B.A.; Nugraha, D.T.; Pásztorné-Huszár, K.; Tóth, A.; Takács, K.; Szerdahelyi, E.; Kiskó, G.; Kovács, Z.; et al. Bioactive Peptides from Liquid Milk Protein Concentrate by Sequential Tryptic and Microbial Hydrolysis. Processes 2021, 9. [Google Scholar] [CrossRef]
- Sossella, F.; Rempel, A.; Monroe Araújo Nunes, J.; Biolchi, G.; Migliavaca, R.; Farezin Antunes, A.C.; Vieira Costa, J.A.; Hemkemeier, M.; Colla, L.M. Effects of Harvesting Spirulina Platensis Biomass Using Coagulants and Electrocoagulation–Flotation on Enzymatic Hydrolysis. Bioresour Technol 2020, 311, 123526. [Google Scholar] [CrossRef]
- Nath, A.; Kailo, G.G.; Mednyánszky, Z.; Kiskó, G.; Csehi, B.; Pásztorné-Huszár, K.; Gerencsér-Berta, R.; Galambos, I.; Pozsgai, E.; Bánvölgyi, S.; et al. Antioxidant and Antibacterial Peptides from Soybean Milk through Enzymatic- and Membrane-Based Technologies. Bioengineering 2020, Vol. 7, Page 5 2019, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- O’Loughlin, I.B.; Murray, B.A.; Brodkorb, A.; FitzGerald, R.J.; Robinson, A.A.; Holton, T.A.; Kelly, P.M. Whey Protein Isolate Polydispersity Affects Enzymatic Hydrolysis Outcomes. Food Chem 2013, 141, 2334–2342. [Google Scholar] [CrossRef]
- Yu, X.X.; Liang, W.Y.; Yin, J.Y.; Zhou, Q.; Chen, D.M.; Zhang, Y.H. Combining Experimental Techniques with Molecular Dynamics to Investigate the Impact of Different Enzymatic Hydrolysis of β-Lactoglobulin on the Antigenicity Reduction. Food Chem 2021, 350. [Google Scholar] [CrossRef]
- Swaminathan, A.V.; Molitor, M.S.; Burrington, K.J.; Otter, D.; Lucey, J.A. Partial Enrichment of Phospholipids by Enzymatic Hydrolysis and Membrane Filtration of Whey Protein Phospholipid Concentrate. JDS Communications 2023, 4, 175–180. [Google Scholar] [CrossRef]
- Cheison, S.C.; Wang, Z.; Xu, S.Y. Hydrolysis of Whey Protein Isolate in a Tangential Flow Filter Membrane Reactor. II. Characterisation for the Fate of the Enzyme by Multivariate Data Analysis. J Memb Sci 2006, 286, 322–332. [Google Scholar] [CrossRef]
- Prieto, C.A.; Guadix, A.; González-Tello, P.; Guadix, E.M. A Cyclic Batch Membrane Reactor for the Hydrolysis of Whey Protein. J Food Eng 2007, 78, 257–265. [Google Scholar] [CrossRef]
- García-Cano, I.; Yeh, P.W.; Rocha-Mendoza, D.; Jiménez-Flores, R. Supercritical CO2 Treatment Reduces the Antigenicity of Buttermilk β-Lactoglobulin and Its Inflammatory Response in Caco-2 Cells. JDS Communications 2021, 2, 1–6. [Google Scholar] [CrossRef]
- Anto, L.; Warykas, S.W.; Torres-Gonzalez, M.; Blesso, C.N. Milk Polar Lipids: Underappreciated Lipids with Emerging Health Benefits. Nutrients 2020, 12. [Google Scholar] [CrossRef]
- Bourlieu, C.; Michalski, M.C. Structure-Function Relationship of the Milk Fat Globule. Curr Opin Clin Nutr Metab Care 2015, 18, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Contarini, G.; Povolo, M. Phospholipids in Milk Fat: Composition, Biological and Technological Significance, and Analytical Strategies. Int J Mol Sci 2013, 14, 2808. [Google Scholar] [CrossRef] [PubMed]
- Rombaut, R.; Dejonckheere, V.; Dewettinck, K. Filtration of Milk Fat Globule Membrane Fragments from Acid Buttermilk Cheese Whey. J Dairy Sci 2007, 90, 1662–1673. [Google Scholar] [CrossRef] [PubMed]
- Fontecha, J.; Brink, L.; Wu, S.; Pouliot, Y.; Visioli, F.; Jiménez-Flores, R. Sources, Production, and Clinical Treatments of Milk Fat Globule Membrane for Infant Nutrition and Well-Being. Nutrients 2020, 12. [Google Scholar] [CrossRef]
- Ries, D.; Ye, A.; Haisman, D.; Singh, H. Antioxidant Properties of Caseins and Whey Proteins in Model Oil-in-Water Emulsions. Int Dairy J 2010, 20, 72–78. [Google Scholar] [CrossRef]
- Spitsberg, V.L.; Ivanov, L.; Shritz, V. Recovery of Milk Fat Globule Membrane (MFGM) from Buttermilk: Effect of Ca-Binding Salts. J Dairy Res 2019, 86, 374–376. [Google Scholar] [CrossRef]
- Calvo, M.V.; Martín-Hernández, M.C.; García-Serrano, A.; Castro-Gómez, M.P.; Alonso-Miravalles, L.; García-Martín, R.; Megino-Tello, J.; Alonso, L.; Fontecha, J. Comprehensive Characterization of Neutral and Polar Lipids of Buttermilk from Different Sources and Its Milk Fat Globule Membrane Isolates. Journal of Food Composition and Analysis 2020, 86, 103386. [Google Scholar] [CrossRef]
- Jakhar, M.; Jain, M. Development of Buttermilk with Fruit Juices and Appraisal of Their Nutritional Qualities. The Pharma Innovation Journal 2019, 8, 404–407. [Google Scholar]
- Barry, K.M.; Dinan, T.G.; Kelly, P.M. Selective Enrichment of Dairy Phospholipids in a Buttermilk Substrate through Investigation of Enzymatic Hydrolysis of Milk Proteins in Conjunction with Ultrafiltration. Int Dairy J 2017, 68, 80–87. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Methods in Food Protein Hydrolysis. In: Enzymic Hydrolysis of Food Protein. Enzymic hydrolysis of food proteins 1986, 110–130. [Google Scholar]
- Perea, A.; Ugalde, U.; Rodriguez, I.; Serra, J.L. Preparation and Characterization of Whey Protein Hydrolysates: Applications in Industrial Whey Bioconversion Processes. Enzyme Microb Technol 1993, 15, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant Activity Applying an Improved ABTS Radical Cation Decolorization Assay. Free Radic Biol Med 1999, 26, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, J.R. Principles of Enzymology for the Food Sciences. Principles of Enzymology for the Food Sciences 2018. [Google Scholar] [CrossRef]
- Beynon, R.J.; Bond, J.S. Proteolytic Enzymes: A Practical Approach. 1989, 259.
- Conway, V.; Gauthier, S.F.; Pouliot, Y. Antioxidant Activities of Buttermilk Proteins, Whey Proteins, and Their Enzymatic Hydrolysates. J Agric Food Chem 2013, 61, 364–372. [Google Scholar] [CrossRef]
- Adamson, N.J.; Reynolds, E.C. Characterization of Casein Phosphopeptides Prepared Using Alcalase: Determination of Enzyme Specificity. Enzyme Microb Technol 1996, 19, 202–207. [Google Scholar] [CrossRef]
- Pessato, T.B.; de Carvalho, N.C.; Tavano, O.L.; Fernandes, L.G.R.; Zollner, R. de L.; Netto, F.M. Whey Protein Isolate Hydrolysates Obtained with Free and Immobilized Alcalase: Characterization and Detection of Residual Allergens. Food Research International 2016, 83, 112–120. [Google Scholar] [CrossRef]
- Saha, B.C.; Hayashi, K. Debittering of Protein Hydrolyzates. Biotechnol Adv 2001, 19, 355–370. [Google Scholar] [CrossRef]
- Motta, J.F.G.; de FREITAS, B.C.B.; de ALMEIDA, A.F.; Martins, G.A. de S.; Borges, S.V. Use of Enzymes in the Food Industry: A Review. Food Science and Technology 2023, 43, e106222. [CrossRef]
- Ceylan, F.D.; Adrar, N.; Günal-Köroǧlu, D.; Subas, B.G.; Capanoglu, E. Combined Neutrase–Alcalase Protein Hydrolysates from Hazelnut Meal, a Potential Functional Food Ingredient. ACS Omega 2022, 8, 1618. [Google Scholar] [CrossRef]
- Doucet, D.; Gauthier, S.F.; Otter, D.E.; Foegeding, E.A. Enzyme-Induced Gelation of Extensively Hydrolysed Whey Proteins by Alcalase: Comparison with the Plastein Reaction and Characterization of Interactions. J Agric Food Chem 2003, 51, 6036–6042. [Google Scholar] [CrossRef]
- Colbert, L.B.; Decker, E.A. Antioxidant Activity of an Ultrafiltration Permeate from Acid Whey. J Food Sci 1991, 56, 1248–1250. [Google Scholar] [CrossRef]
- Mendis, E.; Rajapakse, N.; Kim, S.K. Antioxidant Properties of a Radical-Scavenging Peptide Purified from Enzymatically Prepared Fish Skin Gelatin Hydrolysate. J Agric Food Chem 2005, 53, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.M.P.L.V.O.; Pinho, O.; Mota, M.V.; Tavares, P.; Pereira, A.; Gonçalves, M.P.; Torres, D.; Rocha, C.; Teixeira, J.A. Preparation of Ingredients Containing an ACE-Inhibitory Peptide by Tryptic Hydrolysis of Whey Protein Concentrates. Int Dairy J 2007, 17, 481–487. [Google Scholar] [CrossRef]
- Önay-Uçar, E.; Arda, N.; Pekmez, M.; Yilmaz, A.M.; Böke-Sarikahya, N.; Kirmizigül, S.; Yalçin, A.S. Comparison of Antioxidant Capacity, Protein Profile and Carbohydrate Content of Whey Protein Fractions. Food Chem 2014, 150, 34–40. [Google Scholar] [CrossRef]
- Britten, M.; Lamothe, S.; Robitaille, G. Effect of Cream Treatment on Phospholipids and Protein Recovery in Butter-Making Process. Int J Food Sci Technol 2008, 43, 651–657. [Google Scholar] [CrossRef]
- Wong, P.Y.Y.; Kitts, D.D. Chemistry of Buttermilk Solid Antioxidant Activity. J Dairy Sci 2003, 86, 1541–1547. [Google Scholar] [CrossRef]
- Cheryan, M. Ultrafiltration and Microfiltration Handbook. Ultrafiltration and Microfiltration Handbook 1998. [Google Scholar] [CrossRef]
- Guadix, A.; Camacho, F.; Guadix, E.M. Production of Whey Protein Hydrolysates with Reduced Allergenicity in a Stable Membrane Reactor. J Food Eng 2006, 72, 398–405. [Google Scholar] [CrossRef]
- Leindecker, G.C. Separação Das Proteínas Do Soro Do Leite in Natura Por Ultrafiltração. 2011.
- Atra, R.; Vatai, G.; Bekassy-Molnar, E.; Balint, A. Investigation of Ultra- and Nanofiltration for Utilization of Whey Protein and Lactose. J Food Eng 2005, 67, 325–332. [Google Scholar] [CrossRef]
- Rombaut, R.; Dejonckheere, V.; Dewettinck, K. Filtration of Milk Fat Globule Membrane Fragments from Acid Buttermilk Cheese Whey. J Dairy Sci 2007, 90, 1662–1673. [Google Scholar] [CrossRef]
- Morin, P.; Britten, M.; Jiménez-Flores, R.; Pouliot, Y. Microfiltration of Buttermilk and Washed Cream Buttermilk for Concentration of Milk Fat Globule Membrane Components. J Dairy Sci 2007, 90, 2132–2140. [Google Scholar] [CrossRef] [PubMed]
- Corredig, M.; Roesch, R.R.; Dalgleish, D.G. Production of a Novel Ingredient from Buttermilk. J Dairy Sci 2003, 86, 2744–2750. [Google Scholar] [CrossRef] [PubMed]
- Mulder, H.; Walstra, P. The Milk Fat Globule Emulsion Science as Applied to Milk Products and Comparable Foods; 4th ed.; Commonwealth Agricultural Bureaux: Farnham Royal, 1974; Vol. 19;
- Ali, A.H. Current Knowledge of Buttermilk: Composition, Applications in the Food Industry, Nutritional and Beneficial Health Characteristics. Int J Dairy Technol 2019, 72, 169–182. [Google Scholar] [CrossRef]
- Sabadin, I.S.; Villas-Boas, M.B.; de Lima Zollner, R.; Netto, F.M. Effect of Combined Treatment of Hydrolysis and Polymerization with Transglutaminase on Beta-Lactoglobulin Antigenicity. EUROPEAN FOOD RESEARCH AND TECHNOLOGY 2012, 235, 801. [Google Scholar] [CrossRef]
- Perea, A.; Ugalde, U. Continuous Hydrolysis of Whey Proteins in a Membrane Recycle Reactor. Enzyme Microb Technol 1996, 18, 29–34. [Google Scholar] [CrossRef]
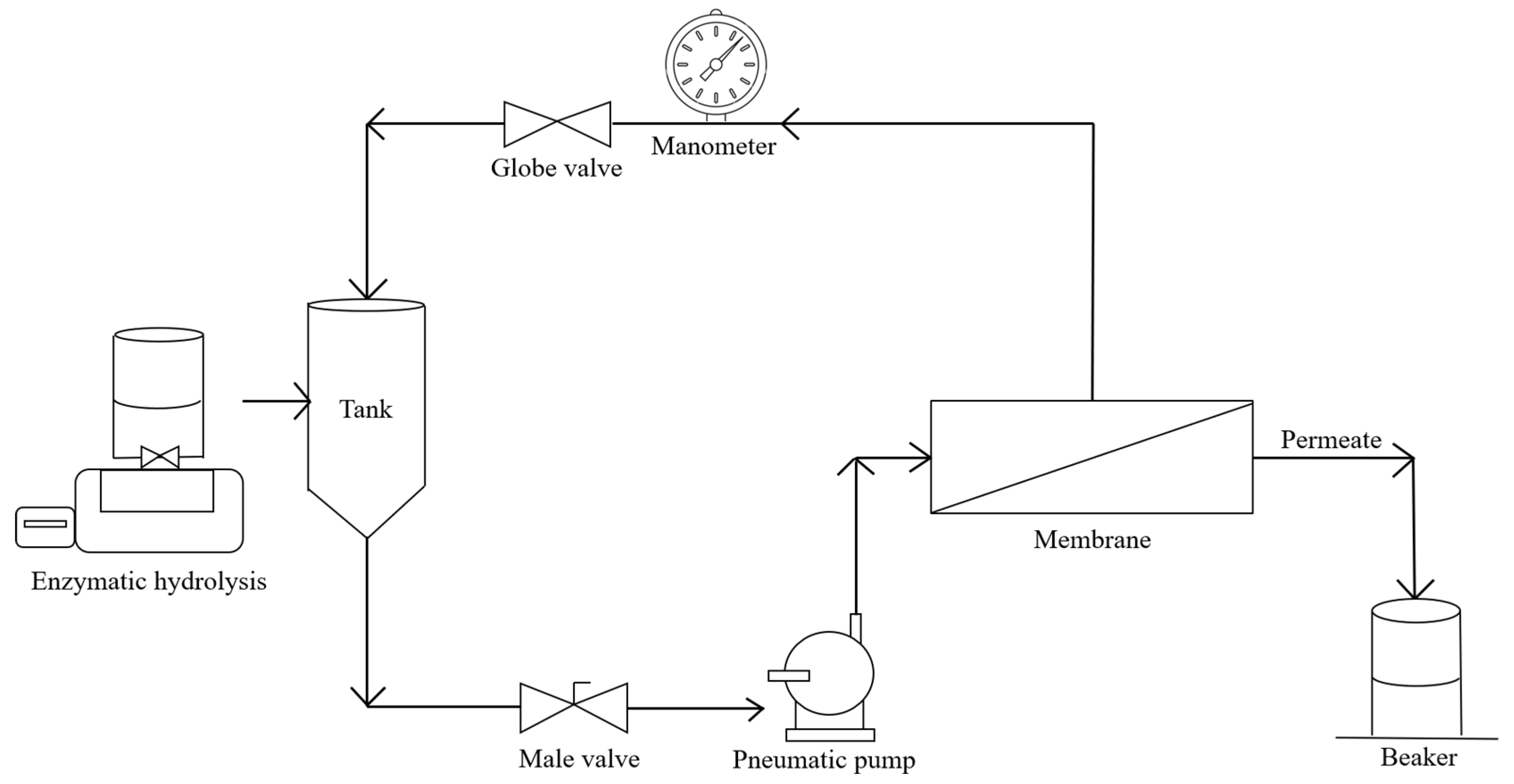
| Enzyme | Action | pH | Temperature (ºC) |
Enzyme concentration (g 100 mL -1) |
Inactivation |
|---|---|---|---|---|---|
| Alcalase® | Endopeptidase | 8.0 | 55 | 0.5 | 90 ºC / 20 min |
| Neutrase® | Metallopeptidase | 7.0 | 55 | 1.25 | 90 ºC / 20 min |
| Prolyve® | Endopeptidase | 8.0 | 55 | 0.5 | 90 ºC / 5 min |
| Sample | Stream | Protein (%) | Lactose (%) | Fats (%) |
|---|---|---|---|---|
|
Buttermilk |
Feed | 2.08±0.03 | 1.76±0.04 | 2.43±0.09 |
| Permeate | 0.93±0.08 a | 0.91±0.09 a | 0.23±1.03 a | |
| Retentate | 1.08±1.08 a | 0.62±1.12 a | 2.12±0.98 a | |
|
Hydrolysed buttermilk |
Feed | 2.08±0.03 | 1.76±0.04 | 2.43±0.09 |
| Permeate | 1.28±0.09 b | 0.94±0.08 a | 0.24±0.08 a | |
| Retentate | 0.78±1.02 b | 0.59±0.09 a | 2.18±0.05 a |
| Enzyme | Time (h) |
Degree of hydrolysis (DH) |
Antioxidant activity ( μmol TE/g protein) |
|---|---|---|---|
| Buttermilk | - | - | 396.81±3.95ª |
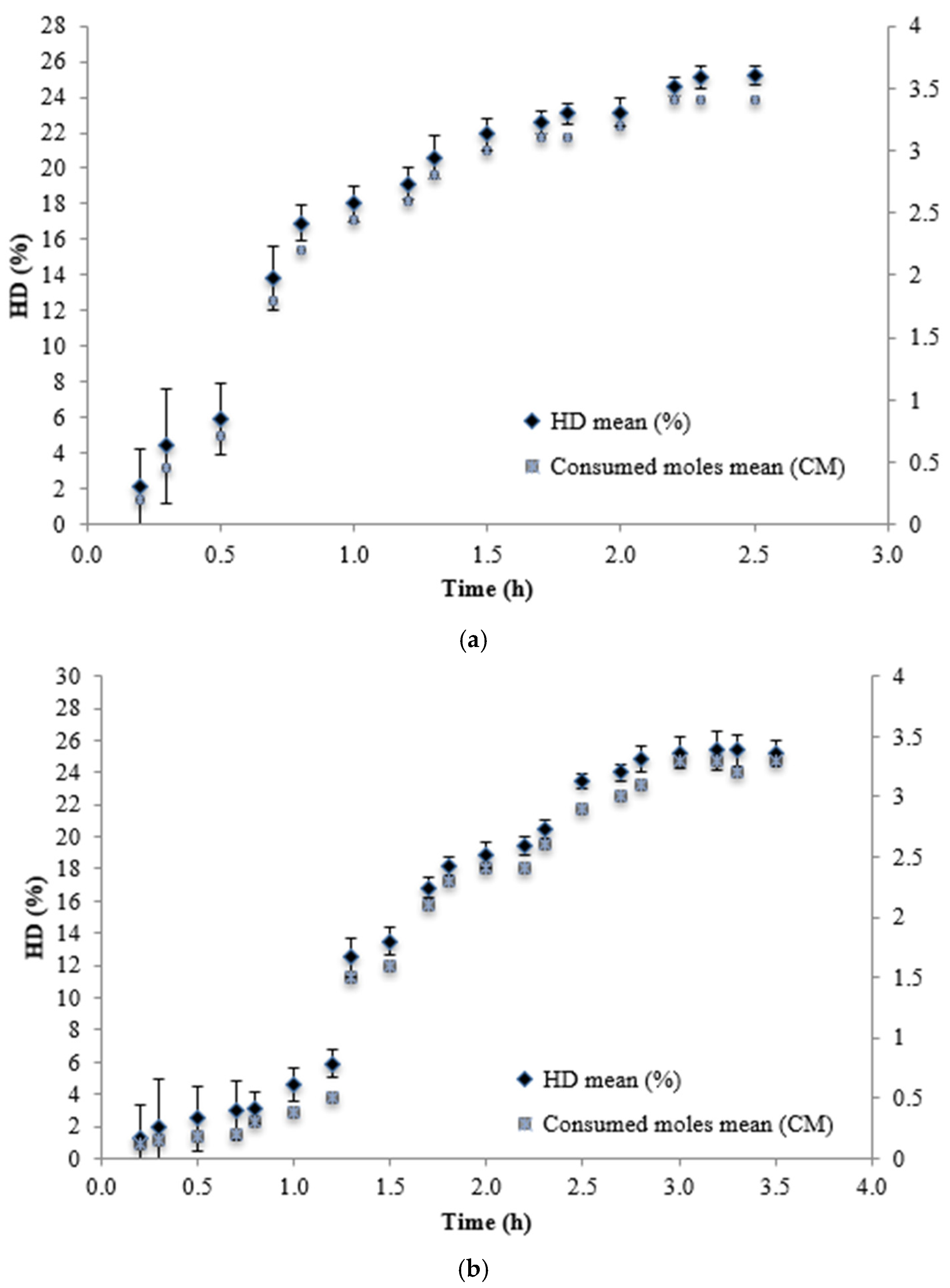
| Alcalase | 2.5 | 25.8 | 803.58±1.57 b |
| Prolyve | 3.5 | 25.2 | 795.73±5.66 b |
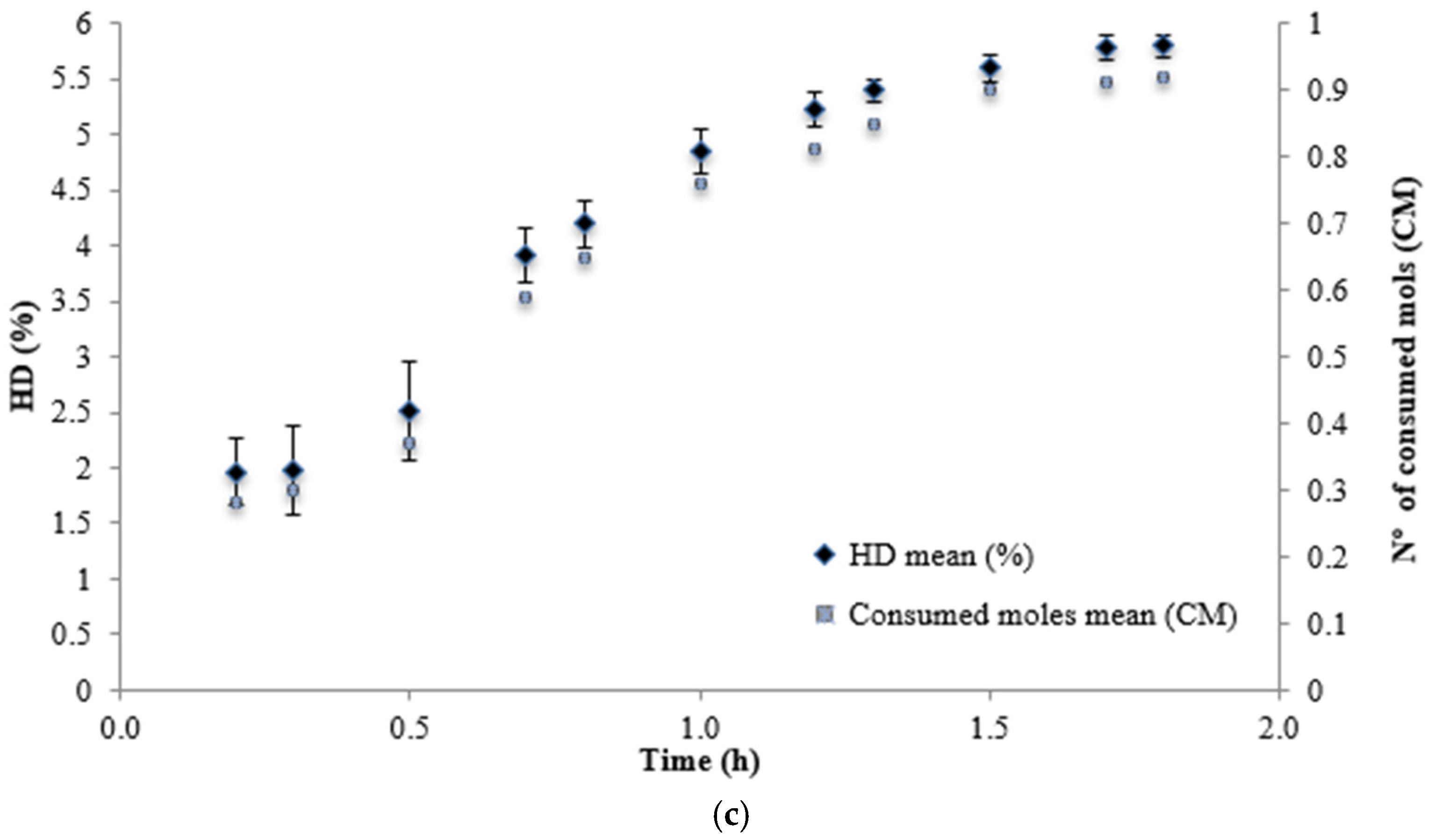
| Neutrase | 1.8 | 5.8 | 418.82±4.16ª |
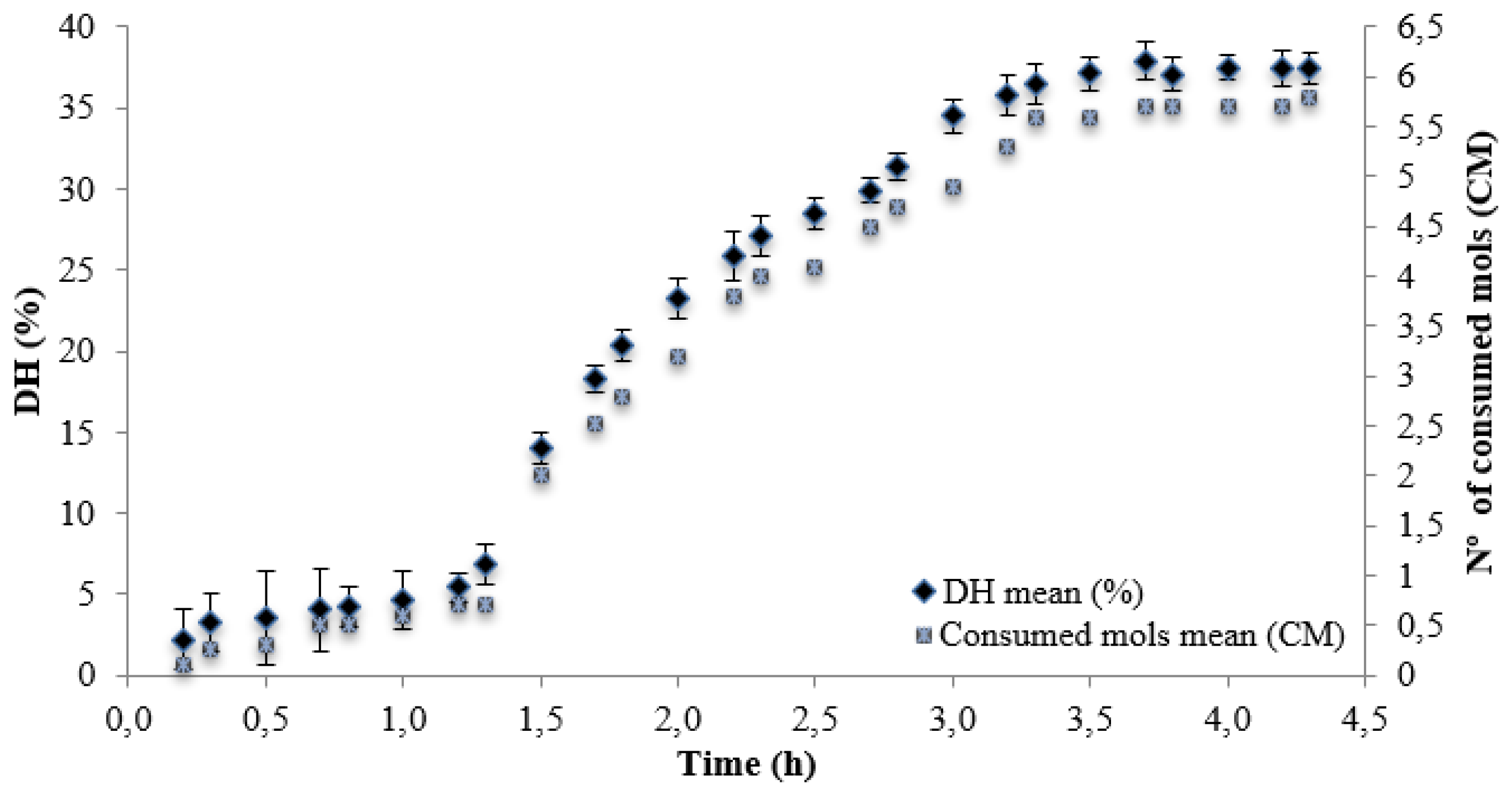
| Alcalase + Neutrase | 4.3 | 34.02 | 834.21±2.40 b |
| Permeate | - | - | 956.87±1.57 c |
| Retentate | - | - | 825.48±3.12 b |
| Enzyme | Time (h) | Hydrolysis Degree (HD) | Average lenght of the chains (No. of residues of aminoacids) |
Average molar mass (kDa) |
|---|---|---|---|---|
| Buttermilk | - | - | 100 | 12,000.00 |
| Alcalase | 2.5 | 25.8 | 4.0 | 483.22 |
| Prolyve | 3.5 | 25.2 | 4.0 | 476.19 |
| Neutrase | 1.8 | 5.8 | 17.2 | 2068.97 |
| Alcalase + Neutrase | 4.3 | 34.02 | 2.9 | 0.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





