1. Introduction
The discovery and development of effective therapies for respiratory viruses such as dengue virus (DENV), influenza, and SARS-CoV-2 is an urgent global health challenge. While traditional antiviral treatments, such as small molecule drugs or antibodies, have been successful in some cases, they rarely have limitations, such as the development of methods and side effects of drug resistance [
1,
2].
Recently, researchers and therapeutic companies have turned their attention to a promising new class of therapeutics, called nanobodies. Nanobodies are small antibody fragments derived from the immune system of camelids and lamas, also known as variable domains of heavy chain antibodies (VHHs), nanobodies have different unique characteristics that make them attractive for the development of antiviral therapies[
3,
4]. These nanobodies are highly stable and can be easily produced in large quantities using bacterium or yeast cells, making them a cost effective option for therapy development [
5]. Moreover, nanobodies have high binding affinity and specificity for their target epitopes or viral antigens, allowing for effective neutralization of viral particles; this approach has shown promising results in preclinical studies, with nanobodies demonstrating strong antiviral activity against broad respiratory viruses such as SARS-CoV-2 [
6,
7].
Nanobodies block viral entry into host cells or inhibit crucial steps in the viral replication cycle by targeting specific viral proteins, such as the spike protein of SARS-CoV-2 or the hemagglutinin protein of influenza virus [
6,
8]. This unique characteristic of nanobodies offers great potential for the fight against respiratory viruses. However, advanced research is needed to understand the specificity of nanobodies and the dynamics between the host and the virus; this opens the door for structural immunology institutions and structural biologists for more investigation and findings. Furthermore, structural experiments of the interactions between respiratory viruses and nanobodies can be offered to optimize their therapeutic potential and develop more effective antiviral treatments [
9]. The structural findings of these interactions will help in the development of nanobodies as potent antiviral therapeutics, offering new hope for controlling and mitigating respiratory viral infections[
10]. Additionally, to nanobodies, other potential approaches for tackling respiratory viruses include the development of safe and effective vaccines targeted at high risk individuals, the transfer of passive immunity through antibody therapies, and the use of antiviral drugs that directly target viral replication processes[
11]. These approaches, combined with current research and collaboration between immunology and virology scientists, hold the key to opening up the power of nanobodies and other innovative diagnostics and therapies to neutralize respiratory viruses and mitigate their impact on global health[
11]. Defined epitopes or recombinant protein arrangements, including the target epitopes, may provide a rational design capable of eliciting convenient humoral or cellular immune responses[
12]. By integrating these strategies with nanobodies, we can enhance the effectiveness and breadth of immune responses against respiratory viruses. Overall, the emergent area of nanobodies shows promising potential for neutralizing respiratory viruses.
2. The Feature of Nanobodies
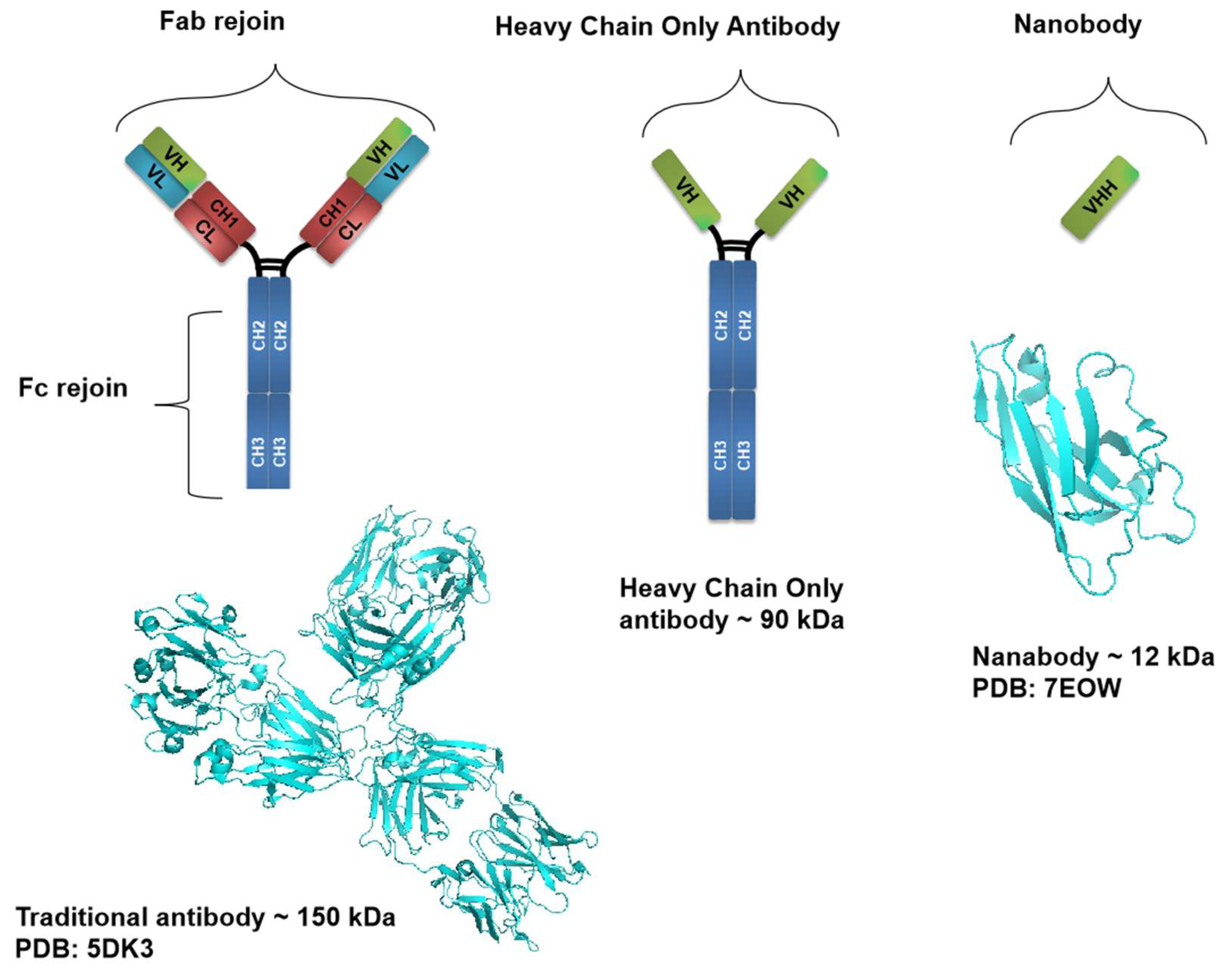
Nanobodies, as small antibody fragments possess unique properties that make them ideal for therapeutic applications for instance, nanobodies have a high specificity and affinity for their viral target antigens, allowing them to neutralize viruses [
13]. The unique structure and small size of nanobodies enable them to reach and bind to viral targets that traditional antibodies cannot access (
Figure 2), they also have high stability and can be produced at a lower cost than conventional antibodies, making them more convenient for diagnosis and therapeutic uses [
5]. Nanobodies are easily engineered to enhance pharmacokinetics and tissue targeting, further improving their therapeutic effectiveness and offering several advantages over traditional antibody therapies for viral respiratory infections [
14]. Additionally, their small size allows them to penetrate the respiratory epithelium more efficiently and reach the infection site, neutralizing the virus particles and inhibiting viral replication [
15]. Furthermore, nanobodies can be produced in large quantities and easily modified to target specific viral protein mutations, epitopes, and viral strains (
Figure 1).
In terms of diagnosis, nanobodies offer several advantages compared to traditional detection methods, including the capability to detect virus particles in real time resulting in rapid results for the management of viral infections and decreasing the side effects[
3]. Nanobodies can be used to detect viruses’ antigens at very low concentrations, resulting in early detection of viral infections even before symptoms apparent, this can be principally crucial in the case of respiratory viruses such as seasonal and pandemic influenza viruses, SARS-CoV-2, and Middle East respiratory syndrome coronavirus (MERS-CoV) where early detection and containment are vital to prevent further spread of the virus[
16].
Revealing the power of nanobodies for detecting and neutralizing respiratory viruses holds huge potential for reforming diagnostics and therapeutics for these viral infections.
3. The Efficacy of Nanobodies Against Respiratory Viruses
Nanobodies can block viral entry into host cells by targeting specific epitopes on the viral surface and inhibiting viral replication, resulting in neutralizing the virus and minimizing the risk of viral escape mutants [
17] table 1. By targeting the N-terminal domain of Spike protein in coronavirus, nanobodies can potentially overcome viral escape and provide a viable target for worldwide vaccines that can protect against different strains of respiratory viruses [
17,
18] (
Figure 2) and table 1. Furthermore, in terms of the neutralization of respiratory viruses, nanobodies have revealed great potential for neutralizing respiratory viruses because of their small size, high stability, and ability to target specific viral proteins. Consequently, nanobodies can be easily produced, modified, and administered, making them promising for the development of antiviral therapeutics, and the combination with advanced imaging techniques and fluorescent sensor molecules can provide valuable insights into infection strategies and role formation of respiratory viruses [
3] table 1. This knowledge can contribute to the development of more effective intervention strategies and the design of targeted therapies against respiratory viral disorders. Moreover, the use of nanobodies in epidemiology and during outbreaks can provide real time for controlling and accurate detection of respiratory viruses in numerous environmental samples, allowing the rapid and sensitive detection of viral particles in water, air, and surfaces[
10]. By using the potential characteristic of nanobodies, we can enhance our ability to detect and monitor respiratory viruses in both clinical and environmental settings, resulting in improving public health responses.
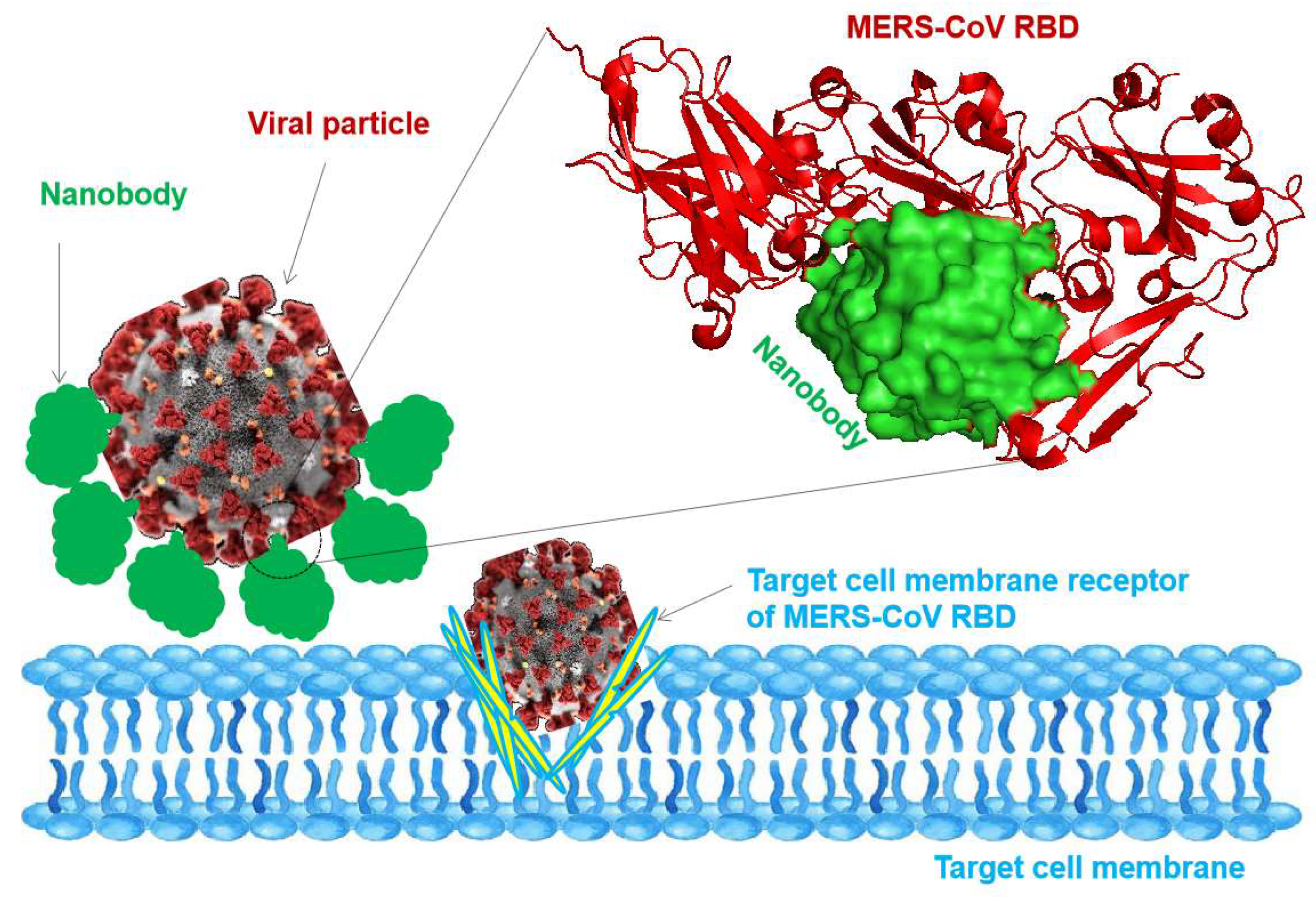
Figure 2.
Blocking and neutralizing of viral particles with nanobodies. The illustration shows the nanobodies molecules inhibiting the viral particle to inter-target cell membrane. The top right shows the blocking interaction between MERS-CoV RBD with a nanobody (PDB: 8YSF).
Figure 2.
Blocking and neutralizing of viral particles with nanobodies. The illustration shows the nanobodies molecules inhibiting the viral particle to inter-target cell membrane. The top right shows the blocking interaction between MERS-CoV RBD with a nanobody (PDB: 8YSF).
Table 1.
Examples of nanobodies targeting respiratory viral proteins, and associated diseases.
Table 1.
Examples of nanobodies targeting respiratory viral proteins, and associated diseases.
| Sr.nu |
Target Viral Protein |
Disease |
Mechanism of Action |
Reference |
| 1 |
Spike RBD |
SARS-CoV-2 |
Blocks receptor binding domain and prevents viral entry |
[17] |
| 2 |
Spike RBD |
SARS-CoV-2 |
Inhibits spike protein conformational change for fusion |
[6] |
| 3 |
Hemagglutinin |
H5N1 |
Prevents hemagglutinin from binding to sialic acid receptors |
[8] |
| 4 |
Hemagglutinin |
H1N1 |
Blocks hemagglutinin from facilitating viral entry |
[9] |
| 5 |
Spike Protein |
MERS-CoV |
Inhibits interaction with DPP4 receptor, blocking entry |
[4] |
| 6 |
Spike Protein |
SARS |
Prevents viral entry by blocking spike protein interaction |
[1] |
| 7 |
F Protein |
RSV |
Stabilizes pre-fusion form, preventing membrane fusion |
[11] |
| 8 |
Spike RBD |
SARS-CoV-2 |
Binds to RBD, blocking ACE2 receptor interaction |
[17] |
| 9 |
Hemagglutinin |
Influenza A |
Inhibits receptor binding, preventing viral entry |
[2] |
| 10 |
Nucleocapsid |
SARS-CoV-2 |
Disrupts nucleocapsid structure, inhibiting replication |
[10] |
| 11 |
Hemagglutinin |
H3N2 |
Blocks hemagglutinin function, preventing viral entry |
[14] |
| 12 |
Spike S2 |
SARS-CoV-2 |
Inhibits spike-mediated membrane fusion |
[15] |
| 13 |
RBD |
MERS-CoV |
Blocks interaction with DPP4 receptor |
[4] |
| 14 |
Spike Protein |
SARS-CoV-2 |
Inhibits viral fusion with host cell membrane |
[6] |
| 15 |
G Protein |
RSV |
Blocks G protein interactions, inhibiting viral entry |
[12] |
| 16 |
ORF7a |
SARS-CoV-2 |
Modulates viral protein function, inhibiting replication |
[13] |
4. Methodologies
4.1. Construction of Nanobody Libraries Using Phage Display Technology
Phage display technology is a powerful method for designing, constructing, and genetically engendered libraries of nanobodies, the process for constructing nanobody libraries using phage display includes the following steps generally [
5,
7,
19]:
- (1)
Separation of lymphocytes: The first step is obtaining the lymphocytes from an immunized camelid or llama with the antigens of interest, typically this is done by drawing blood samples from the animals after the immunization period under ethical committee approval consent.
- (2)
RNA isolation and cDNA synthesis: Using established protocols the total RNA will be extracted from the isolated lymphocytes, and using reverse transcriptase to convert the RNA into complementary DNA (cDNA).
- (3)
Amplification of nanobody sequences: The purified cDNA containing the nanobody sequences will amplify using Polymerase Chain Reaction (PCR) and specific primers to the framework regions of the nanobodies will used to selectively amplify the variable regions of nanobodies.
- (4)
Construction of phage display library: The amplified nanobody sequences are then cloned into a phagemid vector, which contains a gene encoding a coat protein of a bacteriophage, usually M13, phagemid vector confirms that the nanobodies are displayed on the surface of phage particles using molecular biology tool such as sequencing
- (5)
Transformation and phage production: The phagemid vector containing the nanobody library will transform into Escherichia coli (E. coli) cells, and the transformed cells are then grown to a large number of phage particles displaying different nanobodies on the phage surface.
- (6)
Phage selection: The nanobody phage library will be subjected to multiple rounds of affinity selection against the target antigen, usually (2-5 rounds). This involves incubating the library with the antigens of interest and washing away unbound phages several times to obtain a high affinity monoclonal nanobody phage.
- (7)
Elution and amplification: The bound phages will be eluted from the antigen, and the selected nanobody genes will be amply using PCR to generate a new nanobody phage library enriched with nanobodies that have a high affinity for the antigens of interest.
4.2. Bio-Panning Screening of Nanobodies
Bio panning screening technology of nanobodies is used to identify and select specific nanobodies that can be used for different applications[
20]. Bio-panning technology involves several repeated cycles of incubation of a library of nanobodies with varying concentrations of target antigens followed by washing to remove nonspecific nanobodies and eluting the specifically bound nanobodies using trypsin [
5](figure 3). The eluted nanobodies can then be further characterized and evaluated for their binding affinity and specificity, making bio-panning screening a valuable tool in nanobody development and discovery[
5]. Using bio-panning screening, researchers can efficiently identify and select specific nanobodies that bind to a target antigen, resulting in the development of nanobodies with high binding affinity and specificity, making them effective tools for various applications, such as diagnostics, therapeutics, and research[
21,
22].
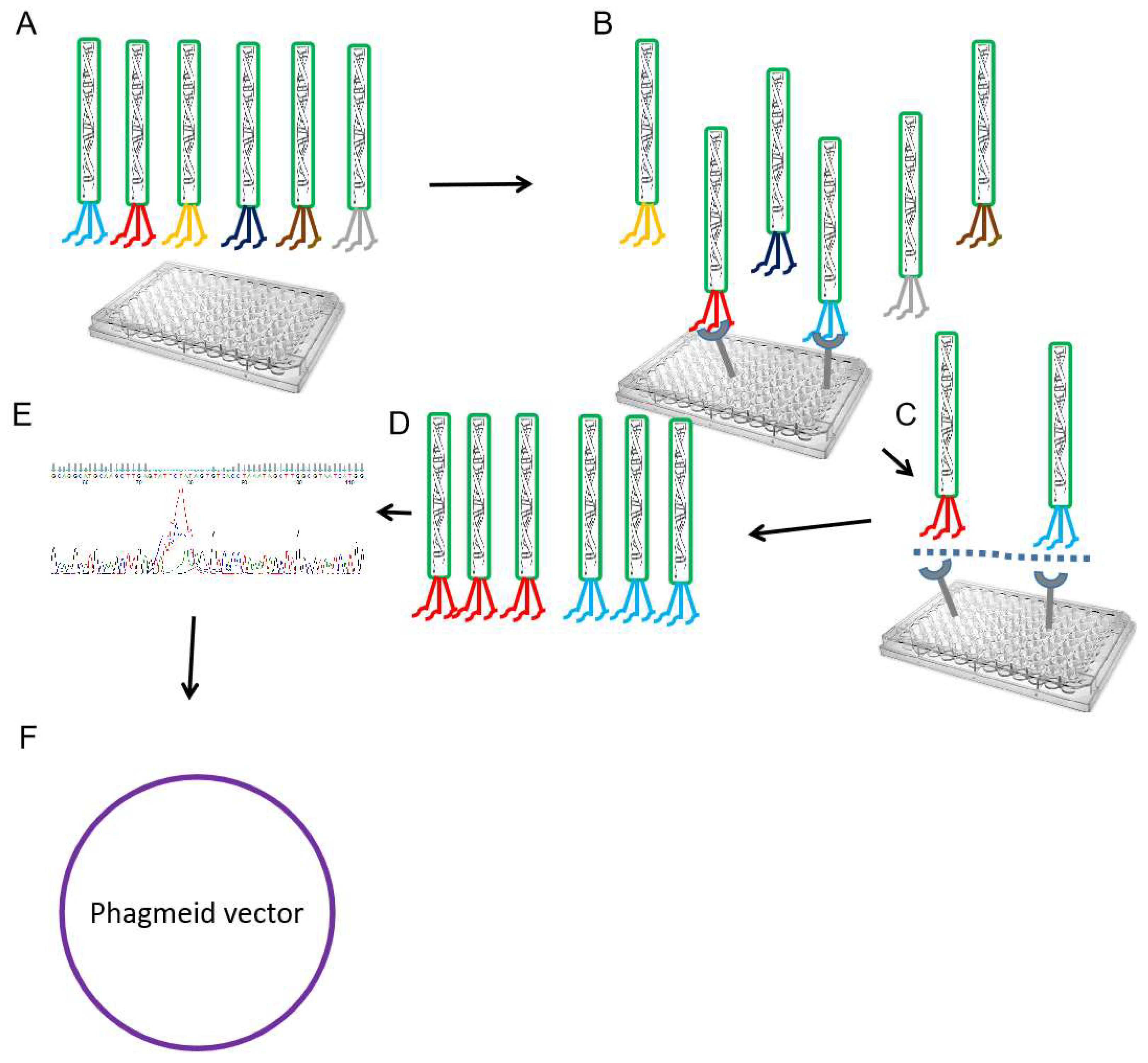
Figure 3.
Nanobody phage display and recombinant phagemid construction. Schematic drawing showing the nanobody phage display selection process (A). Polyclonal phage particles are incubated with antigen-coated ELISA plates (B). Washing to remove unbound polyclonal phage (C). Eluted bound nanobody phage using trypsin to infect bacteria for amplification (D). This cycle of binding, elution, and amplification can be repeated several rounds (3-6) to enrich for phage displaying nanobodies with high affinity (E). Confirmation of selected nanobody phage genes (CDR1-3) sequences through sequencing. (F) Molecular construction of recombinant phagemids using PCR, restriction enzymes, or Gibson Assembly methods. Constructed phagemids can be used for transformation or transfection experiments to produce the selected nanobodies.
Figure 3.
Nanobody phage display and recombinant phagemid construction. Schematic drawing showing the nanobody phage display selection process (A). Polyclonal phage particles are incubated with antigen-coated ELISA plates (B). Washing to remove unbound polyclonal phage (C). Eluted bound nanobody phage using trypsin to infect bacteria for amplification (D). This cycle of binding, elution, and amplification can be repeated several rounds (3-6) to enrich for phage displaying nanobodies with high affinity (E). Confirmation of selected nanobody phage genes (CDR1-3) sequences through sequencing. (F) Molecular construction of recombinant phagemids using PCR, restriction enzymes, or Gibson Assembly methods. Constructed phagemids can be used for transformation or transfection experiments to produce the selected nanobodies.
4.3. Cloning, Expression, and Purification of Selected Nanobodies Based on Viral Antigens
Cloning, expression, and purification of nanobodies based on viral antigens is a multi-step process that includes molecular cloning, protein expression using different expression systems such as bacterial, yeast, mammalian, and insect cells, and purification methods, and the general steps of Cloning, expression, and purification involved [
5,
7]:
Cloning of Nanobody Genes: The selected nanobody genes from the phage nanobodies display library are cloned into expression vectors suitable for protein production such as bacterial plasmid pET30a. These vectors often contain a promoter region to drive gene expression, as well as tags or fusion partners to aid in the purification and detection of the expressed nanobodies. Recently many scientists have engineered to clone the selected nanobody gene to the Fc region of a human IgG1 antibody to increase the affinity and efficacy of the nanobody-Fc antibody against the viral proteins.
Expression of Nanobodies: After the cloning process is successful and confirmed with the PCR and sequencing, cloned expression vectors are transformed or transinfected into a suitable host system for protein expression. Generally used host systems such as bacteria (e.g., E. coli) or yeast (e.g., Saccharomyces cerevisiae), the host cells are then grown for three or four days under suitable conditions to induce the expression of the nanobodies protein.
Purification of Nanobodies: When the nanobodies are expressed, they need to be purified from the host cell lysate or culture supernatant, affinity chromatography is often used, where the nanobodies are selectively bound to a column containing a ligand specific to the tag or fusion partner used during cloning such as Ni-column interacted with the polyhistidine at the vector, expressed with our target nanobodies protein. Other chromatographic techniques, such as ion exchange or size exclusion chromatography, can further purify the nanobodies and remove impurities.
4.4. Characterization and Validation of Nanobodies as Viral Antigens Inhibitors
After the purification of nanobodies as viral antigen inhibitors there are several biochemical and biophysical characterization and validation steps to assess their binding affinity, specificity, and inhibitory potential, and that assay involves the following[
5,
7]:
- (1)
Binding affinity determination: Nanobodies' binding affinity towards the viral antigen is evaluated using biophysical techniques such as surface plasmon resonance (SPR), isothermal titration calorimetry (ITC), size exclusion chromatography (SEC), or enzyme-linked immunosorbent assay (ELISA). These experiments provide information on the strength of the interaction between the nanobody and the viral antigen.
- (2)
Epitope mapping: Epitope mapping studies aim to identify the specific region of the viral antigen recognized by the nanobody. This can be done using techniques such as peptide scanning arrays, alanine scanning mutagenesis, or X-ray crystallography. Understanding the binding site of the nanobody on the viral antigen can help elucidate its inhibitory mechanism.
- (3)
Neutralization assays: To assess the inhibitory potential of nanobodies, neutralization assays are performed. These assays involve exposing the viral pathogen to the nanobody and evaluating its ability to prevent viral entry into host cells or block viral replication. Virus neutralization can be measured through infectivity assays, plaque reduction assays, or viral growth inhibition assays.
- (4)
In vivo validation: Nanobodies showing promising inhibitory effects in vitro can be further validated in animal models. Animal studies, such as murine models or non-human primates, can provide insights into the nanobody's efficacy, safety, pharmacokinetics, and immunogenicity.
- (5)
Structural characterization: Structural studies, such as X-ray crystallography or cryo-electron microscopy, can be conducted to determine the atomic level details of the nanobody viral antigen complex. These studies provide insights into the binding mode and conformational changes occurring upon complex formation.
- (6)
Optimization and engineering: Nanobodies can be further optimized and engineered to enhance their binding affinity, stability, or half-life. Technologies like phage display based affinity maturation, site-directed mutagenesis, or antibody humanization can be applied to improve the nanobody's properties for therapeutic applications.
5. Conclusion and Future Directions
Nanobodies showed a revolutionary advancement in the antiviral therapies field, offering exclusive advantages over conventional antibodies, their small size, high stability, and cost-effective production methods make them ideal candidates for addressing the global health challenges of respiratory viruses. As research progresses, several key areas must be explored to unlock the full potential of nanobodies in the fight against viral infections. Continued structural optimization through advanced techniques like cryo-electron microscopy (cryo-EM) and X-ray crystallography will extend our understanding of nanobody viral antigen interactions, guiding the design of more effective nanobodies with enhanced binding affinities and specificity. Furthermore, developing nanobodies that target conserved regions across various viral strains could lead to broad spectrum antiviral therapies, providing robust defenses against emerging variants. Exploring the synergistic effects of nanobodies in combination with traditional antiviral drugs, vaccines, or other immunotherapeutic agents will be crucial in enhancing treatment efficacy and reducing the risk of viral resistance. Transitioning from preclinical studies to clinical trials is essential for validating the safety and efficacy of nanobodies in humans, and establishing optimal dosage routines, delivery methods, and long-term effects. Furthermore, leveraging the unique properties of nanobodies for rapid, sensitive diagnostic tests could revolutionize the early detection of viral infections. Additionally, the development of testing kits using nanobody based detection methods has the potential to enhance public health response mechanisms significantly. In summary, the integration of structural insights, engineering innovations, and clinical applications will be essential in advancing nanobodies as effective antiviral therapeutics. The collaboration between immunologists, virologists, and structural biologists will be essential in ensuring the successful implementation of nanobody based therapies in clinical settings, ultimately contributing to improved public health outcomes worldwide.
Author Contributions
AM, conceptualized the main idea, extracted the data, wrote the original draft, and formatted the manuscript for submission. MM, reviewed and formatted the manuscript for submission. All authors read and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Acknowledgements
The authors acknowledge the Deanship of Scientific Research at Biotech Department at Omdurman Islamic University, for the supportive cooperation.
Conflicts of Interest
None.
References
- Sharma, A., et al., Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): a global pandemic and treatment strategies. International journal of antimicrobial agents, 2020. 56(2): p. 106054.
- Meganck, R.M. and R.S. Baric, Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases. Nature medicine, 2021. 27(3): p. 401-410. [CrossRef]
- Steeland, S., R.E. Vandenbroucke, and C. Libert, Nanobodies as therapeutics: big opportunities for small antibodies. Drug discovery today, 2016. 21(7): p. 1076-1113. [CrossRef]
- Sroga, P., D. Safronetz, and D.R. Stein, Nanobodies: a new approach for the diagnosis and treatment of viral infectious diseases. Future Virology, 2020. 15(3): p. 195-205. [CrossRef]
- Mohammed, A., et al., Generation, biochemical characterizations and validation of potent nanobodies derived from alpaca specific for human receptor of advanced glycation end product. Biochemical and Biophysical Research Communications, 2021. 581: p. 38-45. [CrossRef]
- Ma, H., et al., Potent neutralization of SARS-CoV-2 by hetero-bivalent alpaca nanobodies targeting the spike receptor-binding domain. Journal of virology, 2021. 95(10): p. 10.1128/jvi. 02438-20. doi:10.1128/JVI.
- Ma, H., et al., Hetero-bivalent nanobodies provide broad-spectrum protection against SARS-CoV-2 variants of concern including Omicron. Cell Research, 2022. 32(9): p. 831-842.
- Ibanez, L.I., et al., Nanobodies with in vitro neutralizing activity protect mice against H5N1 influenza virus infection. Journal of Infectious Diseases, 2011. 203(8): p. 1063-1072. [CrossRef]
- Skehel, J.J. and M.D. Waterfield, Studies on the primary structure of the influenza virus hemagglutinin. Proceedings of the National Academy of Sciences, 1975. 72(1): p. 93-97. [CrossRef]
- Kamat, S., M. Kumari, and C. Jayabaskaran, Nano-engineered tools in the diagnosis, therapeutics, prevention, and mitigation of SARS-CoV-2. Journal of Controlled Release, 2021. 338: p. 813-836. [CrossRef]
- Pantaleo, G., et al., Antibodies to combat viral infections: development strategies and progress. Nature Reviews Drug Discovery, 2022. 21(9): p. 676-696. [CrossRef]
- Alizadeh, M., et al., Designing a novel multi-epitope vaccine against Ebola virus using reverse vaccinology approach. Scientific reports, 2022. 12(1): p. 7757. [CrossRef]
- Bhattacharya, M., et al., Therapeutic applications of nanobodies against SARS-CoV-2 and other viral infections: Current update. International Journal of Biological Macromolecules, 2023. 229: p. 70-80.
- Gao, J., M. Gui, and Y. Xiang, Structural intermediates in the low pH-induced transition of influenza hemagglutinin. PLoS Pathogens, 2020. 16(11): p. e1009062.
- Chen, X., et al., A cell-free nanobody engineering platform rapidly generates SARS-CoV-2 neutralizing nanobodies. Nature communications, 2021. 12(1): p. 5506.
- Minatel, V.M., et al., Nanobodies: a promising approach to treatment of viral diseases. Frontiers in Immunology, 2024. 14: p. 1303353. [CrossRef]
- Huo, J., et al., Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nature structural & molecular biology, 2020. 27(9): p. 846-854.
- Esparza, T.J., et al., High affinity nanobodies block SARS-CoV-2 spike receptor binding domain interaction with human angiotensin converting enzyme. Scientific Reports, 2020. 10(1): p. 22370.
- Chen, Y.-L., et al., Research progresses and applications of fluorescent protein antibodies: a review focusing on nanobodies. International Journal of Molecular Sciences, 2023. 24(5): p. 4307. [CrossRef]
- Veugelen, S., et al., Screening and characterization strategies for nanobodies targeting membrane proteins. Methods in enzymology, 2017. 584: p. 59-97.
- Lu, Q., et al., Development of multivalent nanobodies blocking SARS-CoV-2 infection by targeting RBD of spike protein. Journal of nanobiotechnology, 2021. 19: p. 1-12.
- Li, J.-F., et al., Generation and characterization of a nanobody against SARS-CoV. Virologica Sinica, 2021. 36(6): p. 1484-1491.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).