Submitted:
05 December 2024
Posted:
06 December 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Regression with Interaction
3. Meta Analysis for the Main Effect and Interaction Effect
3.1. Fixed effect meta-analysis
3.2. Random-effect meta-analysis
4. Covariance in Meta-Analysis
4.1. Relationship Between Covariance and Correlation Coefficient r
4.2. Fixed-Effect Meta-Analysis for Correlation Coefficient r
4.3. Random-Effect Meta-Analysis for Correlation Coefficient r
5. Example: Analyzing the 10 Cohort Studies in Table 1
5.1. Meta-Analysis for
5.2. Meta-Analysis for
5.3. Meta-Analysis for r
5.4. Overall Covaraince for and
- Fixed-effect model:
- Random-effect model:
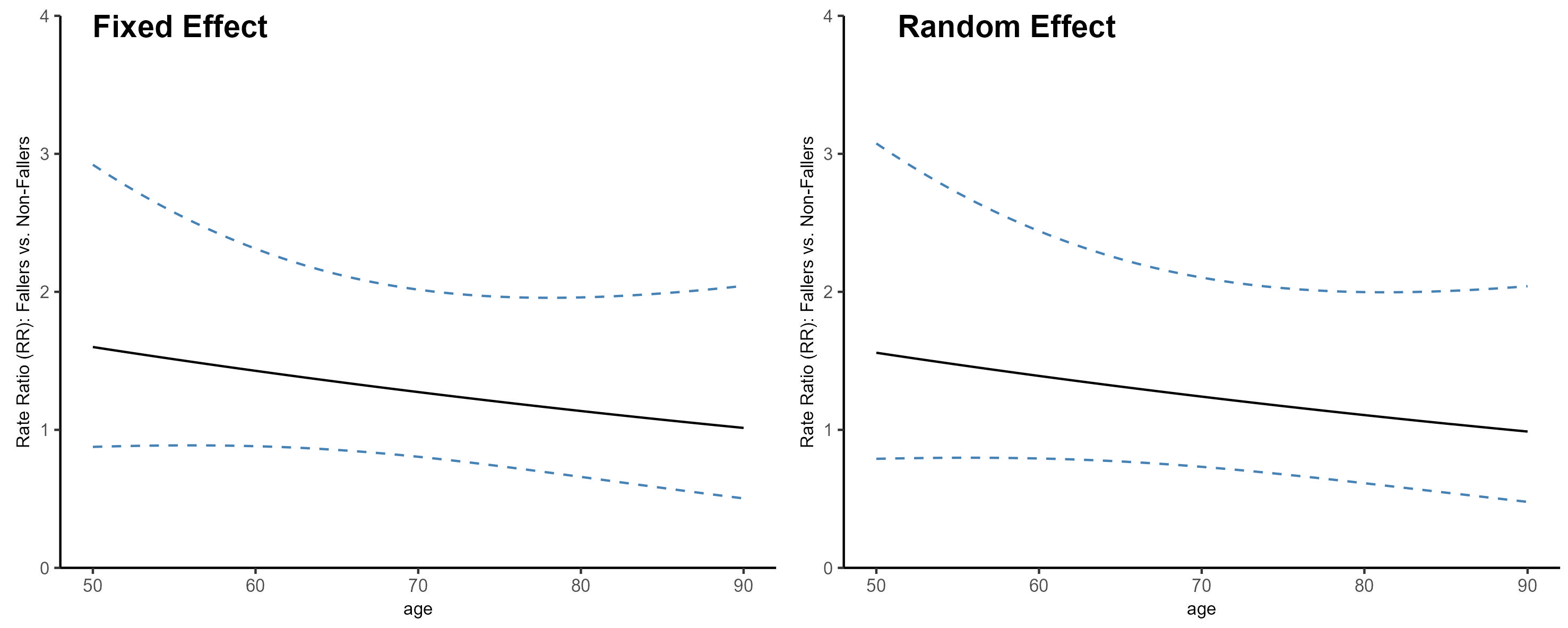
5.5. Calculating the Dffect of Falls on Hip Fracture at Different Ages and Its 95% Confidence Intervals
6. An R Package for Calculating Overall Covariance in Meta-Analysis
7. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liesbeth Vandenput, Helena Johansson, Eugene V. McCloskey. et al. A meta-analysis of previous falls and subsequent fracture risk in cohort studies. Osteoporos International 2024, 35, 469–494. [Google Scholar] [CrossRef]
- John A Kanis, Helena Johansson , Eugene V McCloskey, et al. Previous fracture and subsequent fracture risk: a meta-analysis to update FRAX. Osteoporos International 2023, 34, 2027–2045. [Google Scholar] [CrossRef]
- Danielle L. Burke, Joie Ensor, Richard D. Riley. Burke, Joie Ensor, Richard D. Riley. Meta-analysis using individual participant data: one-stage and two-stage approaches, and why they may differ. Statistics in Medicine 2017, 36, 855–87. [Google Scholar] [CrossRef]
- Richard D Riley, Joie Ensor, Miriam Hattle, et al. Two-stage or not two-stage? That is the question for IPD meta-analysis projects. Research Synthesis Methods 2023, 14, 903–910. [Google Scholar] [CrossRef]
- Adolfo Figueiras, Jose Maria Domenech-Massons, Carmen Cadarso. Regression models: calculating the confidence interval of effects in the presence of interactions. Statistics in medicine 1998, 17, 2099–2105. [Google Scholar] [CrossRef]
- Shahjahan Khan. Meta-Analysis Methods for Health and Experimental Studies; Springer, 2020.
- Michael Borenstein, Larry V.Hedges, Julian P.T.Higgins, HannahR.Rothstein. Introduction to meta-analysis; John Wiley & Sons, 2021.
- Guido Schwarzer, James R. Carpenter , Gerta Rucke, Meta-Analysis with R; Springer, 2015.
- Rebecca DerSimonian, Nan Laird. Meta-analysis in clinical trials. Controlled clinical trials 1986, 7, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Agustin Garcia Asuero, Ana Sayago, Gustavo Gonzlez. The correlation coefficient: An overview. Critical reviews in analytical chemistry 2006, 36, 41–59. [Google Scholar] [CrossRef]
- Guido Schwarze. meta: An R package for meta-analysis. R news 2007, 7, 40–45. [Google Scholar]
- Wolfgang Viechtbauer. Conducting meta-analyses in R with the metafor package. Journal of statistical software 2010, 36, 1–48. [Google Scholar]
- Milos Brankovic, Isabella Kardys, Ewout W Steyerberg, et al. Understanding of interaction (subgroup) analysis in clinical trials. European journal of clinical investigation 2019, 49, e13145. [Google Scholar] [CrossRef] [PubMed]
- Rui Wang, Stephen W Lagakos, James H Ware, et al. Statistics in medicine—reporting of subgroup analyses in clinical trials. New England Journal of Medicine 2007, 357, 2189–2194. [Google Scholar] [CrossRef] [PubMed]
- Marty Richardson, Paul Garner, and Sarah Donegan. Interpretation of subgroup analyses in systematic reviews: a tutorial. Clinical Epidemiology and Global Health 2019, 7, 192–198. [Google Scholar] [CrossRef]
- Christopher H Schmid, Paul C Stark, Jesse A Berlin, et al. Meta-regression detected associations between heterogeneous treatment effects and study-level, but not patient-level, factors. Journal of clinical epidemiology 2004, 57, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Loukia M. Spinelia, Nikolaos Pandis. Spinelia, Nikolaos Pandis. Problems and pitfalls in subgroup analysis and meta-regression. American journal of orthodontics and dentofacial orthopedics 2020, 158, 901–904. [Google Scholar] [CrossRef]
- Pim Cuijpers, Jason W Griffin, Toshi A Furukawa. The lack of statistical power of subgroup analyses in meta-analyses: a cautionary note. Epidemiology and Psychiatric Sciences 2021, 30, e78. [Google Scholar] [CrossRef] [PubMed]
- The Stata news, In the spotlight: Multivariate meta-analysis, Available online: https://www.stata.com/stata-news/news37-1/multivariate-meta-analysis/ (accessed on 28 November 2024).
- Sheng Luo, Yong Chen, Xiao Su, Haitao Chu. mmeta: an R package for multivariate meta-analysis. Journal of Statistical Software 2014, 56, 1–26. [Google Scholar]
- Richard D Riley, Malcolm J Price, Dan Jackson, et al. Multivariate meta-analysis using individual participant data. Research synthesis methods 2015, 6, 157–174. [Google Scholar] [CrossRef]
- Dimitris Mavridis, Georgia Salanti. A practical introduction to multivariate meta-analysis. Statistical methods in medical research 2013, 22, 133–158. [Google Scholar] [CrossRef] [PubMed]
- Dan Jackson, Richard Riley, Ian R. White. Multivariate meta-analysis: potential and promise. Statistics in medicine 2011, 30, 2481–2498. [Google Scholar] [CrossRef]
- Antonio Gasparrini, Benedict Armstrong, Michael G Kenward. Multivariate meta-analysis for non-linear and other multi-parameter associations. Statistics in Medicine 2012, 31, 3821–3839. [Google Scholar] [CrossRef]
- Andy P. Field. Field. Meta-analysis of correlation coefficients: a Monte Carlo comparison of fixed-and random-effects methods. Psychological methods 2001, 6, 161–180. [Google Scholar] [CrossRef] [PubMed]
| Cohort | var() | var() | cov() | |||
|---|---|---|---|---|---|---|
| A | 3.0014 | -0.0240 | 2.9419 | 0.0005 | -0.0333 | 5000 |
| B | 1.1488 | -0.0677 | 14.6165 | 0.0029 | -0.2000 | 30000 |
| C | 1.5819 | -0.0936 | 15.8097 | 0.0022 | -0.1825 | 10000 |
| D | 2.0349 | -0.0139 | 3.6954 | 0.0318 | -0.3230 | 3000 |
| E | -4.1219 | 0.0225 | 5.2448 | 0.0009 | -0.0629 | 2000 |
| F | 1.2506 | -0.0020 | 11.2628 | 0.002 | -0.144 | 5000 |
| G | 2.3383 | -0.0173 | 5.2458 | 0.0009 | -0.0644 | 1000 |
| H | -3.1343 | 0.0483 | 9.0698 | 0.0013 | -0.103 | 2000 |
| I | 1.3066 | -0.0005 | 13.7763 | 0.0031 | -0.2000 | 800 |
| J | 3.7753 | -0.021 | 15.0422 | 0.0019 | -0.1635 | 3200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





