1. Introduction
Histone deacetylases (HDACs) are crucial in the epigenetic regulation of DNA replication and transcription. They act on histone proteins within the cell nucleus, contributing to nucleosome formation and regulating chromatin folding and extension. HDACs remove acetyl groups from lysine residues on histone proteins, promoting chromatin compaction, while histone acetyltransferases (HATs) acetylate lysine residues, leading to chromatin relaxation. Chromatin extension makes DNA accessible to enzymes involved in replication and transcription, whereas chromatin folding restricts access to these enzymes [
1].
HDAC involvement has been linked to numerous diseases, suggesting that HDAC inhibitors (HDACi) could be explored as potential antitumor and neuroprotective agents. Recent studies have shown HDAC involvement in neurodegenerative disorders such as Alzheimer’s disease [
2] as well as in various cancers [
3].
The search for effective HDACi has led to the development and commercialization of drugs like Belinostat, Panobinostat, and Vorinostat [
4], all of which contain the hydroxamic acid functional group, a well-recognized zinc-binding group (ZBG) [
5].
This underscores the need to optimize molecular docking protocols to enhance the identification of potentially active HDACi, particularly during virtual screening efforts. However, accurately predicting the binding free energies and poses of HDAC inhibitors has been challenging due to the involvement of zinc ions, which are essential for the catalytic activity of HDACs. As a divalent metal cation, zinc has unique coordination and electrostatic properties that can complicate docking and scoring procedures. One of the main challenges with docking software is the inadequate parameterization of zinc ions, which are often treated as simple divalent cations. This ignores zinc’s ability to form coordination complexes.
Accurately determining binding free energies for ligands targeting metalloproteins, particularly those containing zinc ions, necessitates specialized computational approaches [
6]. Standard docking programs often fail to model metal center interactions effectively, leading to inaccurate predictions. To address this limitation, advanced computational tools and methods have been developed specifically to model metalloproteins, focusing on zinc coordination environments. Examples of these tools, listed in chronological order of implementation, include FlexX [
7,
8,
9,
10], AutoDockZn [
11], MpsDockZn [
12], GM-DockZn [
13], Autodock-Vina [
14,
15], GPDOCK [
16], AutoDock Bias [
17] (a modification within the AutoDock suite), and MetalDock [
18].
For zinc-containing HDAC enzymes, the following methodology can be employed to enhance docking accuracy and binding energy calculations: i) preprocessing the zinc coordination sphere by explicitly modeling its coordination geometry (typically tetrahedral) to ensure accurate geometry during docking setup. This includes specifying coordination partners such as histidine, aspartate, water molecules, and potential ligand interactions with the zinc ion. ii) Precise ligand parameterization is essential, given that accurate protonation states and charge distribution are crucial for HDAC inhibitors, particularly for functional groups like hydroxamates or carboxylates, which interact with zinc.
We employed the AutoDock Bias tool to implement the first step, which introduces scoring biases favoring hydrogen bond donors near the zinc ion, thereby better mimicking zinc coordination bonds. Recently, the authors proposed an advanced approach known as “metalloprotein bias docking” (MBD), which extends the AutoDock Bias method [
19]. MBD has shown superior accuracy in predicting poses and ligand-binding free energies, although this approach has yet to be applied explicitly to HDACs.
For the second step, we assessed whether to parameterize hydroxamic acids in their
O-undissociated or
O-dissociated forms, given that most studies use the undissociated state [
6,
20]. Since 2007, several authors have demonstrated, through DFT computational studies, that hydroxamic acid tends to be deprotonated within the active site [
21,
22,
23]. However, to the best of our knowledge, only a few studies, primarily by the Sippl group, have specifically investigated this dissociated form. These studies provide limited explanation beyond its potential to more accurately reproduce the native bidentate chelation poses observed in co-crystallized ligand structures [
24,
25,
26,
27,
28,
29,
30].
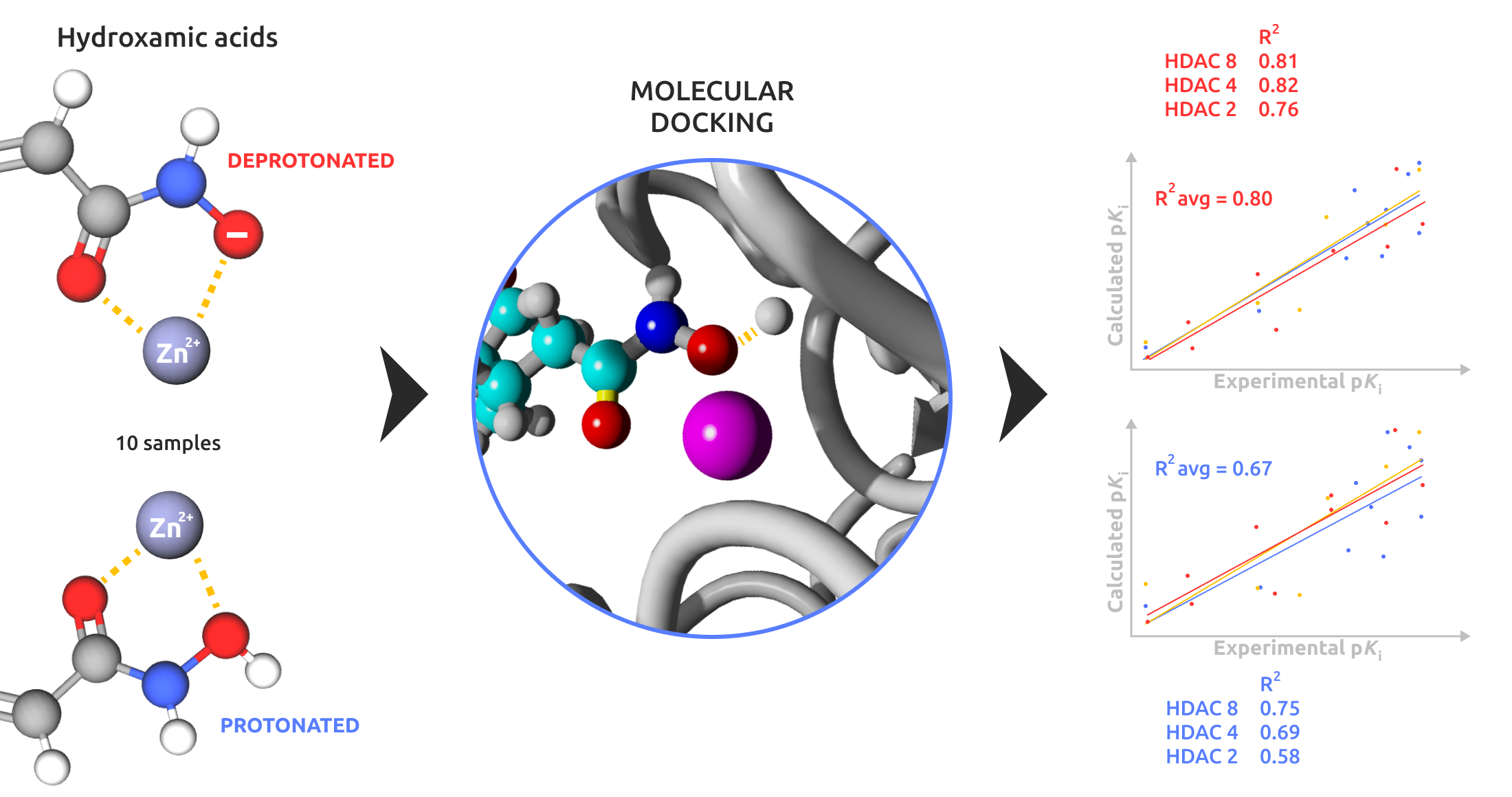
Considering that HDAC isoforms typically exhibit conserved interactions near the zinc-binding site, involving histidine and tyrosine residues that stabilize the zinc-binding group (ZBG) through hydrogen bonds [
31], we examined two possible protonation states, shown in
Figure 1 for HDAC 8.
We focused on the two histidines adjacent to the hydroxamic acid moiety, hypothesizing that the deprotonated form would exhibit stronger metal ion chelation due to increased negative charge density on the hydroxamic oxygen, facilitated by the histidine residues in proximity [
32]. The primary objective of this study was to evaluate whether a docking algorithm optimized for zinc interactions could distinguish between protonated and deprotonated hydroxamic acid ligands, identifying the best poses and calculated binding energies to match experimental values closely. This evaluation would support rapid virtual screening for large ligand libraries, optimizing expected results.
2. Results and Discussion
2.1. Coordination Geometry of the Zinc Ion
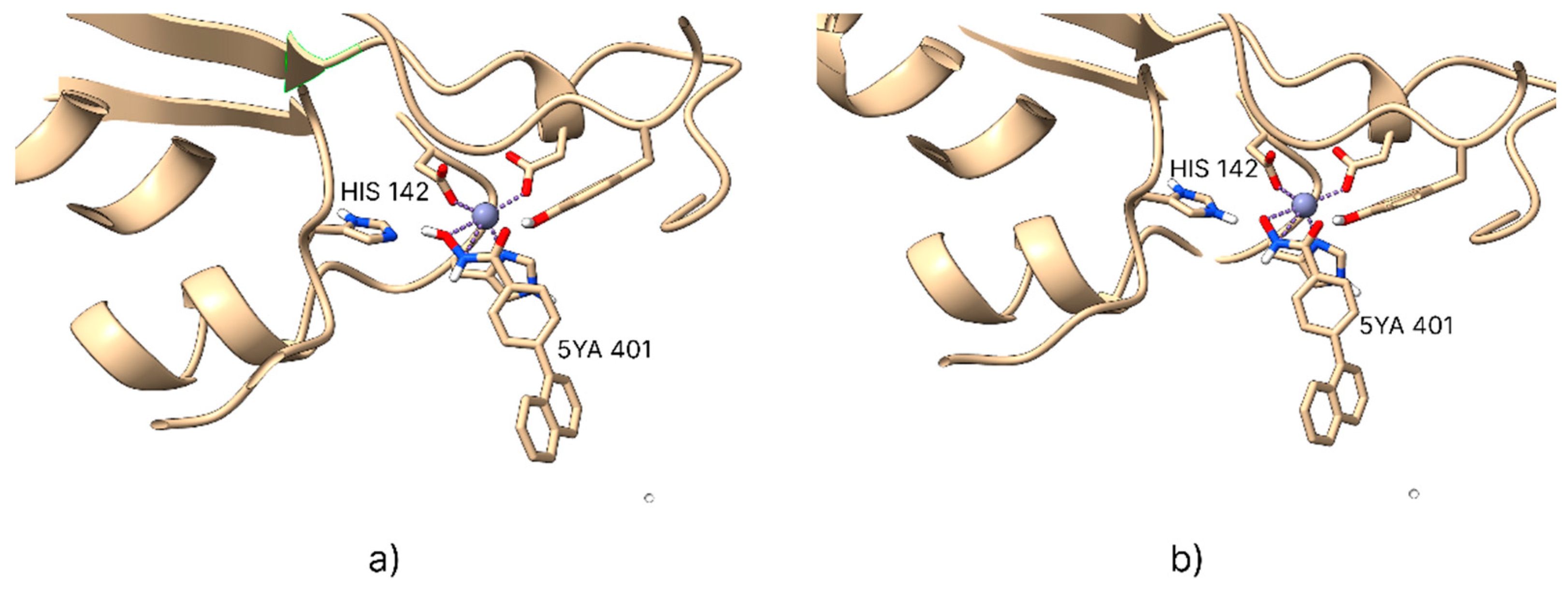
A notable limitation of the AutoDock Bias algorithm is its inability to assign biases to the zinc ion due to the complex nature of its coordination bonds, which cannot be generalized and must be assessed individually for each protein. To address this, the coordination geometry of the zinc ion within HDAC enzymes was studied using co-crystallized PDB structures containing hydroxamic acid as a co-crystallized ligand. The analysis revealed that the coordination geometry is consistently maintained across all examined PDB entries, with zinc ions coordinated by two histidine and one aspartate residues. Based on this observation, a Python script was developed to predict the ideal positions of hydrogen bond acceptor groups in the ligand, using the known coordination geometry once the co-crystallized ligand is removed.
This approach accurately predicts the ideal interaction positions of hydroxamic ligands, enabling the calculation of bias positions for each PDB structure even when hydroxamic acid is not present as the co-crystallized ligand (
Figure 2).
2.2. Active Ligands Selection
To ensure biological consistency, active ligands for each HDAC isoform were selected from the same study [
21]. Importantly, all the activity values, expressed as
Ki, were derived using the same binding assay, facilitating accurate comparison across datasets. Using activity data from different and/or biologically incompatible assays could compromise experimental integrity, potentially skewing results and undermining the reliability of subsequent docking studies. Each ligand was manually analyzed and categorized into one of three distinct initial datasets (
Table 1) corresponding to a specific HDAC isoform. Furthermore, ligands were selected with diverse selectivity profiles wherever feasible across the three isoforms to further substantiate the rigor of the molecular selection process.
2.3. Introduction of Biases
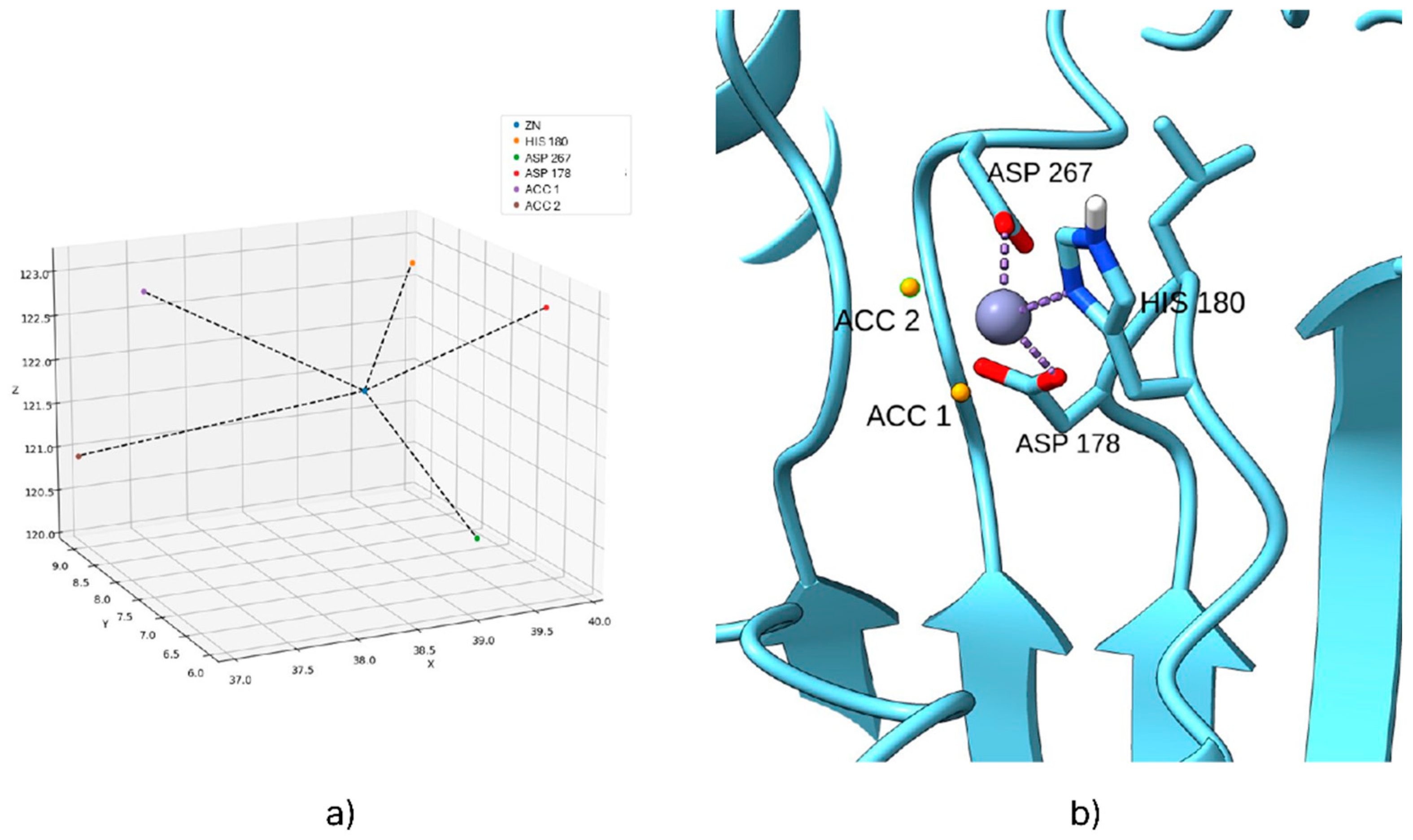
Biases were introduced after thoroughly analyzing the binding site for each HDAC isoform. The most critical interactions between the co-crystallized ligand and the target protein were identified, and specific biases related to the zinc atom were applied to these interactions. One significant limitation of AutoDock Bias is its inability to parameterize the zinc atom accurately. To overcome this, a Python script was developed to predict the biases to be applied for zinc ion interactions within HDAC enzymes.
The correct interaction geometry between zinc ions and hydroxamic acids was studied using multiple co-crystallized structures containing hydroxamic acids and ligands. These studies confirmed that the zinc coordination geometry remains consistent across various co-crystals, with the zinc ion always coordinated by one histidine residue and two aspartate residues. Among the aspartate residues, one typically positions slightly further from the zinc ion than the other. The Python script takes this coordination geometry into account, calculating the ideal interaction coordinates between the hydroxamic ligand and the zinc ion based on the relative positions of the histidine and aspartate residues in the PDB structure (with the ligand removed).
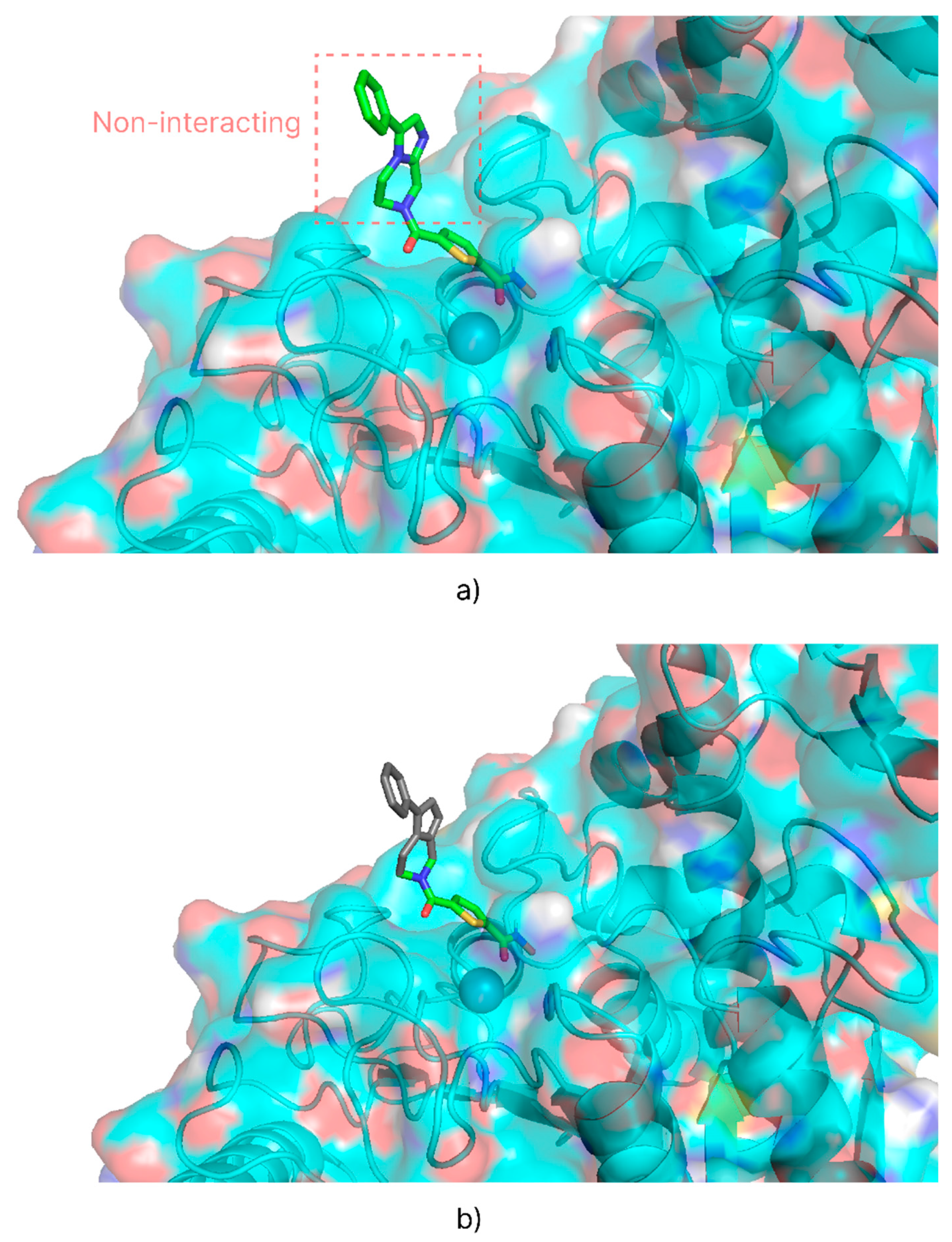
Root mean square deviation (RMSD) studies were performed using the methods described in the AutoDock Bias documentation to confirm the validity of the introduced biases. Re-docking of the co-crystallized ligand was performed first without and then with the introduction of the biases. Biases that maintained unchanged or improved the RMSD value were considered valid. For the RMSD analysis of the HDAC 4 isoform, only the portion of the ligand that interacts with the target (
Figure 3, b) was considered, neglecting the part that escapes from the catalytic site (
Figure 3, a). Since the latter has no intermolecular interactions with the protein’s amino acids, it was considered insignificant to the evaluation file.
The results (
Table 2) confirm the validity of the introduced biases because all the RMSD values obtained by introducing the biases fall within the acceptability limit of 3 Å, usually considered a good RMSD value. Specifically, the analyses performed for HDAC isoforms 4 and 8 confirm the quality of the re-docking of the co-crystallized ligand, while for HDAC isoform 2, there is a substantial improvement due to the introduction of the biases.
The Python script, a tutorial on predicting the bias positions for zinc ions in HDAC enzymes, grid position coordinates, biases coordinates, and all relevant virtual screening (VS) files are available in our GitHub repository (see SI).
2.4. Virtual Screening
VSs of ligand sets prepared in both protonated and deprotonated forms were performed. The free energy of binding (ΔG) associated with the first pose was used to evaluate the interaction of each ligand. To ensure reproducibility, energy values were extracted from the output files by considering the “Mean Binding Energy” of the most populated cluster as the representative energy value. In cases where clusters had similar populations, the free energy of binding was averaged across these clusters.
The derived Δ
G value was then converted to the corresponding
Ki value using the thermodynamic relationship:
The equation was solved at T = 310 K, with R = 0.0019872036 kcal/K×mol. Subsequently, the Ki values were transformed into their negative logarithm, pKi.
The so calculated pKi values were then compared with experimental pKi values via a scatter plot, and a regression analysis was conducted to obtain the correlation coefficient (R2), which was used to evaluate and compare the results.
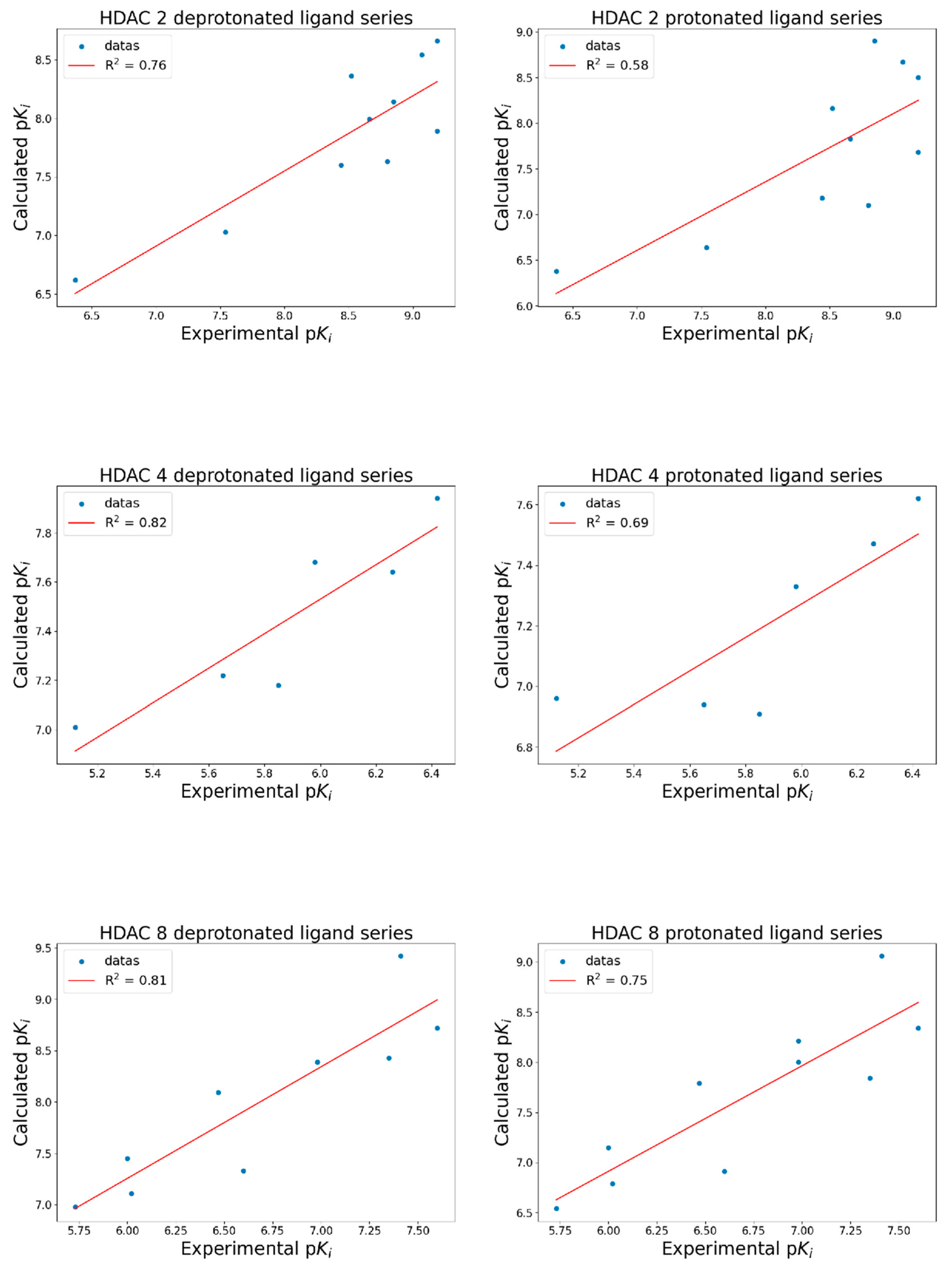
The docking results (
Figure 4) indicate that deprotonated hydroxamic ligands correlate better with experimental data than protonated ligands. The correlation between deprotonated ligands and experimental data consistently outperformed protonated ligands.
Specifically, the R2 for deprotonated versus protonated ligands were as follows: for HDAC 2, R2deprotonates = 0.76 and R2protonates = 0.58; for HDAC 4, R2deprotonates = 0.82 and R2protonates = 0.69; for HDAC 8, R2deprotonates = 0.81 and R2protonates = 0.75.
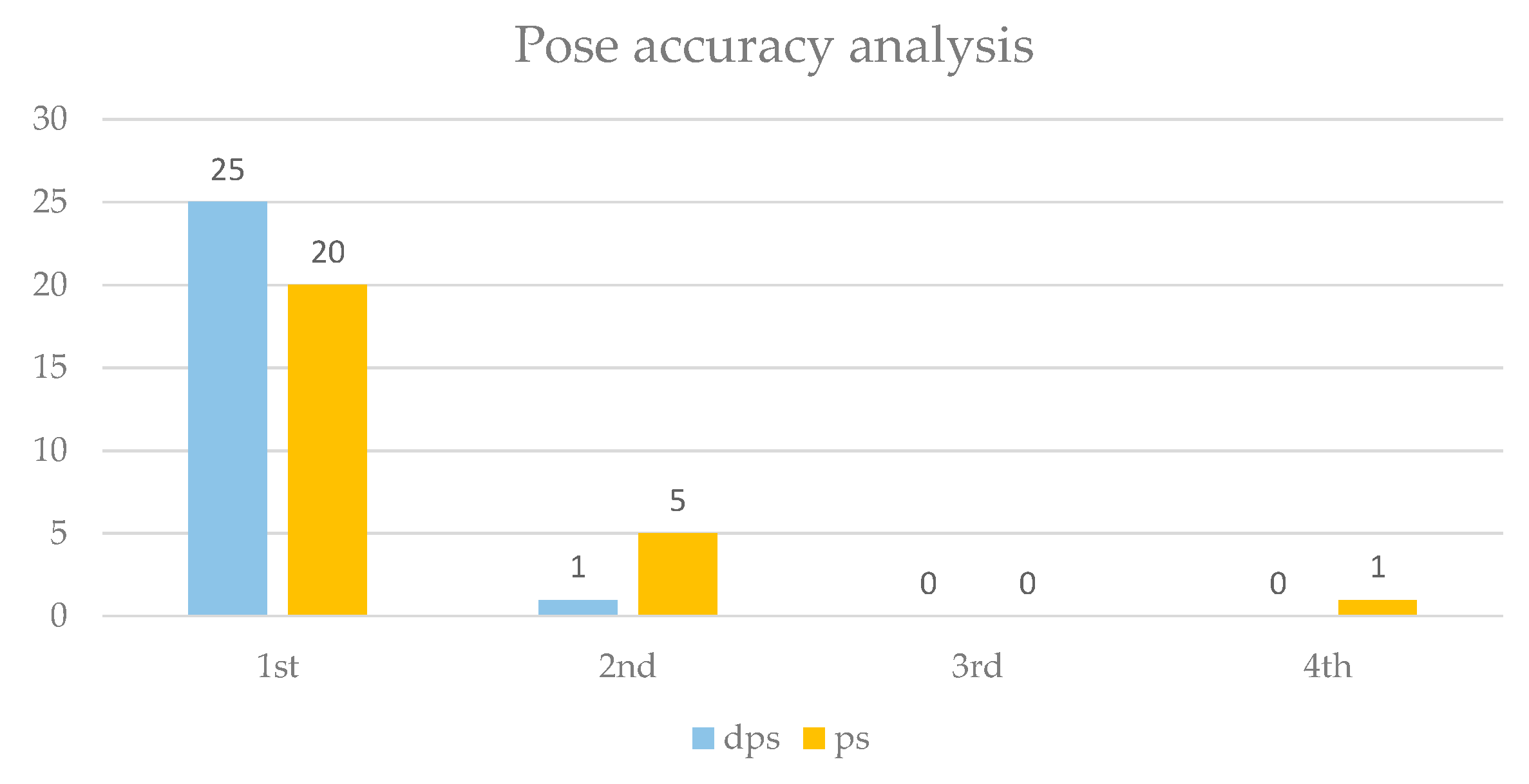
In addition, the accuracy of the poses generated by the docking algorithm was analyzed, focusing on the ability to correctly position the hydroxamic group of the ligand to coordinate the zinc ion. Each target’s selected poses (
Tables S1 and S2) were considered. The chosen poses were those in which the ligand’s hydroxamic group demonstrated coordination binding with the zinc ion. 26 poses were analyzed across the protonated and deprotonated ligand series. The number of instances where the corrected pose, with proper coordination of the hydroxamic group, appeared in the first, second, third, or fourth position in the docking results was recorded to evaluate pose accuracy. Percent pose accuracy was calculated as the ratio of the instances where the correct pose ranked first to the total poses analyzed. As illustrated in
Figure 5, deprotonated ligands displayed a significantly higher accuracy, with the hydroxamic group correctly positioned in the first pose in 96.2% of cases, compared to only 76.9% for protonated ligands. This represents a 25.1% improvement in the prediction of correct poses when using deprotonated ligands, highlighting their enhanced suitability for accurate docking predictions.
Two key findings emerge from this study. The most significant result is the comparison between protonated and deprotonated ligands. As predicted theoretically, the interaction between the zinc ion and the electron lone pairs is more favorable when the electron density on the electronegative atom is higher, as in the case of the deprotonated ligand. Experimental results strongly support this theoretical assumption. Specifically, the docking algorithm generated energy values that correlated better with experimental data for the deprotonated ligand series than the protonated ones. These findings underscore two crucial factors for enhancing the accuracy of ΔG of binding predictions in metalloprotein docking, particularly for HDACs. First, ligands should be deprotonated, and the histidine residues in the proteins should be protonated. Second, the custom zinc coordination Python script we developed for AutoDock Bias should be used when working with HDACs to ensure the zinc interaction is appropriately accounted for by the docking algorithm.
The second noteworthy result is comparing the quality of the poses obtained. By using hydroxamic ligands in the deprotonated form, the docking algorithm can more easily identify the group coordinating the zinc ion, almost always positioning it as the best result. This further supports the conclusion that deprotonated ligands are preferable, showing greater consistency with experimental data.
3. Materials and Methods
3.1. Biological Data and 3D Structures Generation
The chemical structures of ligands active on HDAC enzymes were selected from a single source to ensure biological consistency across binding assays. Hydroxamic acid-containing ligands were prioritized, and their affinity values (
Ki) were converted from µM to nM, subsequently expressed as decimal negative logarithms (p
Ki = −log
Ki). Ligand preparation was conducted first using RDKit open-source toolkit for cheminformatics (v2023.09.6) using the Python script rdconf.py download from GitHub repository “rdkit-scripts” (
https://github.com/dkoes/rdkit-scripts), through which where SMILES codes were used to generate 3D structures in single conformer and using ETKDG knowledge-based method [
33] instead of distance geometry.
Ligand preparation continued in Open Babel (v. 3.1.1) [
34], where sdf format was converted into pdbqt format for compatibility with AutoDock4 [
35], reflecting physiological pH (7.4) states. For
O-deprotonated ligands, the hydroxamic OH hydrogen atom was manually removed before 3D structure generation, after which structures were optimized as outlined.
3.2. Protein Preparation
The protein structures were downloaded from the Protein Data Bank (
https://www.rcsb.org/). Due to their significant pharmaceutical importance, human-HDAC isoforms 2, 4, and 8 (PDB IDs: 4LXZ, 2VQM, and 5FCW, respectively) were chosen. Chain A was selected for analysis for each protein structure.
Proteins were prepared for docking using the ChimeraX software (v. 1.7.1) [
36]. Non-relevant domains, water molecules, and other non-functional co-crystallized molecules for docking were removed. The preparation of the proteins in the presence of the co-crystallized ligand was carried out using the integrated dockprep option in the software, which performs protonation at physiological pH, amino acid discretization according to appropriate rotamers, and assignment of charges to metal ions (in our case, Zn
2+).
The prepared structures were minimized using the YASARA software (v. 23.12.24), force field AMBER14 was applied, and minimization was done according to the default settings via the Energy Minimization option.
Subsequently, the ligand was removed, and the protein’s three-dimensional structure was saved in pdb format compatible with the AutoDock4 algorithm.
To docking ligands in the deprotonated hydroxamate form, the above structures of each HDAC isoform were generated with the His145 (HDAC 2), His158 (HDAC 4), and His142 (HDAC 8) in the protonated HIP form.
3.3. Molecular Docking Simulation
Virtual screening was performed through the AutoDock Bias suite integrated within the AutoDock Tools (v. 1.5.7). The algorithm employed was AutoDock4 (v. 4.2.6) with the Lamarckian Genetic Algorithm (LGA). Through AutoGrid4 (v. 4.2.6), the grid was centered with the co-crystallized ligand, and its size was extended by 70 grid points in three dimensions of space. Docking was performed with the GA parameters configured as follows: ga_runs = 100, ga_pop_size = 150, ga_num_evals = 2500000, ga_num_generations = 27000, ga_elitism = 1, ga_mutation_rate = 0.02, ga_crossover_rate = 0.8, ga_crossover_mode = two points, ga_cauchy_alpha = 0.0, ga_cauchy_beta = 1.0, number of generations for picking worst individual = 10.
Before docking, all the scripts given in AutoDock Bias’s ‘User guide’ were used to prepare the ligand, receptor, AutoGrid Grid Parameter File, and all docking parameter files. Grid position coordinates used for docking and bias coordinates applied to each target were reported in
Tables S3 and S4.
4. Conclusions
In conclusion, this study evaluated the efficacy of a zinc-optimized docking algorithm in distinguishing between protonated and deprotonated states of hydroxamic acid ligands when targeting HDAC enzymes. By rigorously analyzing both protonation states, we aimed to determine which form yielded docking poses and binding energy predictions that more closely aligned with experimental data. Our findings indicate that deprotonated ligands consistently demonstrated a stronger correlation with experimental results across all HDAC targets. Specifically, for HDAC 2, 4, and 8, deprotonated ligands showed superior R² values, highlighting the enhanced accuracy of docking predictions with the deprotonated form.
This outcome underscores the importance of considering ligand protonation states in molecular docking studies of zinc-containing enzymes, as using deprotonated hydroxamic groups in silico offers docking energy values that more reliably mirror experimental outcomes. Ultimately, these findings provide a foundation for improved computational workflows, facilitating rapid virtual screening for HDAC inhibitors and other metalloprotein-targeting drugs, thereby streamlining the discovery of high-affinity, zinc-binding ligands with significant therapeutic potential.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org. Table S1: Complete docking data for deprotonated ligand series. The Δ
G values shown were derived by considering the Mean Binding Energy of the first ligand pose in which the hydroxamic functional group coordinates the zinc ion; Table S2: Complete docking data for protonated ligand series. The Δ
G values shown were derived by considering the Mean Binding Energy of the first ligand pose in which the hydroxamic functional group coordinates the zinc ion; Table S3: Grid position coordinates used for docking. For all targets, grid spacing was set to 0.375 Å, and the number of grid points in dimensions (x, y, z) was set to 70×70×70; Table S4: Bias coordinates applied to each target. A comprehensive, step-by-step tutorial detailing the methodology and providing all experimental data to ensure a thorough understanding of the methodology and facilitate the reproducibility of the results is available on GitHub:
https://github.com/rocco-b/Zinc_Package.
Author Contributions
Conceptualization, A.R. and R.B.; methodology, A.R. and R.B.; software, A.R. and R.B.; validation, A.R., R.B., A.C. and E.A.; formal analysis, R.B. and L.P.; investigation, A.R., R.B. and A.C.; resources, A.R., R.B., L.P., E.A. and A.M.; data curation, R.B. and A.C.; writing—original draft preparation, A.R., R.B. and A.C.; writing—review and editing, A.R., R.B., A.C., L.P., E.M. and A.M.; visualization, R.B. and A.C.; supervision, A.R.; project administration, A.R., A.C., and A.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Acknowledgments
This work was supported by i) the Italian Ministry of Health, Piano di Sviluppo e Coesione del Ministero della Salute 2014-2020, Project: Pharma-HUB - Hub per il riposizionamento di farmaci nelle malattie rare del sistema nervoso in età pediatrica (CUP E63C22001680001 - ID T4-AN-04), ii) Programma di ricerca CN00000013 “National Centre for HPC, Big Data and Quantum Computing”, finanziato dal Decreto Direttoriale di concessione del finanziamento n.1031 del 17.06.2022 a valere sulle risorse del PNRR MUR – M4C2 – Investimento 1.4 - Avviso “Centri Nazionali” - D.D. n. 3138 del 16 dicembre 2021, iii) Italian MUR, PRIN-2022 PNRR program (Project code: P20224L3NK, CUP E53D23015810001, PE5) funded by the European Union – Next Generation EU, (D.D. 1409 del 14/09/2022 MUR), and iv) Italian MUR, PRIN-2022, Prot. 2022Z3BBPE_006, PE5.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Milazzo, G.; Mercatelli, D.; Di Muzio, G.; Triboli, L.; De Rosa, P.; Perini, G.; Giorgi, F.M. Histone Deacetylases (HDACs): Evolution, Specificity, Role in Transcriptional Complexes, and Pharmacological Actionability. Genes 2020, 11, 556. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, R.; Wang, G.; Zhang, Y. The Development Prospection of HDAC Inhibitors as a Potential Therapeutic Direction in Alzheimer’s Disease. Transl Neurodegener 2017, 6, 19. [Google Scholar] [CrossRef]
- Ramaiah, M.J.; Tangutur, A.D.; Manyam, R.R. Epigenetic Modulation and Understanding of HDAC Inhibitors in Cancer Therapy. Life Sciences 2021, 277, 119504. [Google Scholar] [CrossRef]
- Yoon, S.; Eom, G.H. HDAC and HDAC Inhibitor: From Cancer to Cardiovascular Diseases. Chonnam Med J 2016, 52, 1. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, J.; Jiang, Q.; Zhang, L.; Song, W. Zinc Binding Groups for Histone Deacetylase Inhibitors. Journal of Enzyme Inhibition and Medicinal Chemistry 2018, 33, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Curcio, A.; Rocca, R.; Alcaro, S.; Artese, A. The Histone Deacetylase Family: Structural Features and Application of Combined Computational Methods. Pharmaceuticals 2024, 17, 620. [Google Scholar] [CrossRef] [PubMed]
- Warren, G.L.; Andrews, C.W.; Capelli, A.-M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. A Critical Assessment of Docking Programs and Scoring Functions. J. Med. Chem. 2006, 49, 5912–5931. [Google Scholar] [CrossRef] [PubMed]
- Rarey, M.; Kramer, B.; Lengauer, T.; Klebe, G. A Fast Flexible Docking Method Using an Incremental Construction Algorithm. Journal of Molecular Biology 1996, 261, 470–489. [Google Scholar] [CrossRef] [PubMed]
- Kubinyi, H. Success Stories of Computer-Aided Design. In Computer Applications in Pharmaceutical Research and Development; Ekins, S., Ed.; Wiley, 2006; pp. 377–424 ISBN 978-0-471-73779-7.
- Gastreich, M.; Lilienthal, M.; Briem, H.; Claussen, H. Ultrafast de Novo Docking Combining Pharmacophores and Combinatorics. J Comput Aided Mol Des 2007, 20, 717–734. [Google Scholar] [CrossRef] [PubMed]
- Santos-Martins, D.; Forli, S.; Ramos, M.J.; Olson, A.J. AutoDock4 Zn : An Improved AutoDock Force Field for Small-Molecule Docking to Zinc Metalloproteins. J. Chem. Inf. Model. 2014, 54, 2371–2379. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.; Liao, S.; Gu, J.; Jiang, H.; Wang, X.; Li, H. An Accurate Metalloprotein-Specific Scoring Function and Molecular Docking Program Devised by a Dynamic Sampling and Iteration Optimization Strategy. J. Chem. Inf. Model. 2015, 55, 833–847. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Lyu, N.; Diao, H.; Jin, S.; Zeng, T.; Zhou, Y.; Wu, R. GM-DockZn: A Geometry Matching-Based Docking Algorithm for Zinc Proteins. Bioinformatics 2020, 36, 4004–4011. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J Comput Chem 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Wang, K. GPDOCK: Highly Accurate Docking Strategy for Metalloproteins Based on Geometric Probability. Briefings in Bioinformatics 2023, 24, bbac620. [Google Scholar] [CrossRef]
- Arcon, J.P.; Modenutti, C.P.; Avendaño, D.; Lopez, E.D.; Defelipe, L.A.; Ambrosio, F.A.; Turjanski, A.G.; Forli, S.; Marti, M.A. AutoDock Bias: Improving Binding Mode Prediction and Virtual Screening Using Known Protein–Ligand Interactions. Bioinformatics 2019, 35, 3836–3838. [Google Scholar] [CrossRef] [PubMed]
- Hakkennes, M.L.A.; Buda, F.; Bonnet, S. MetalDock: An Open Access Docking Tool for Easy and Reproducible Docking of Metal Complexes. J. Chem. Inf. Model. 2023, 63, 7816–7825. [Google Scholar] [CrossRef] [PubMed]
- Clemente, C.M.; Prieto, J.M.; Martí, M. Unlocking Precision Docking for Metalloproteins. J. Chem. Inf. Model. 2024, 64, 1581–1592. [Google Scholar] [CrossRef]
- Drakontaeidi, A.; Pontiki, E. A Review on Molecular Docking on HDAC Isoforms: Novel Tool for Designing Selective Inhibitors. Pharmaceuticals (Basel) 2023, 16, 1639. [Google Scholar] [CrossRef] [PubMed]
- Bradner, J.E.; West, N.; Grachan, M.L.; Greenberg, E.F.; Haggarty, S.J.; Warnow, T.; Mazitschek, R. Chemical Phylogenetics of Histone Deacetylases. Nat Chem Biol 2010, 6, 238–243. [Google Scholar] [CrossRef]
- Wang, D.; Helquist, P.; Wiest, O. Zinc Binding in HDAC Inhibitors: A DFT Study. J. Org. Chem. 2007, 72, 5446–5449. [Google Scholar] [CrossRef] [PubMed]
- Ruibo, W.; Zhenyu, L.; Zexing, C.; Yingkai, Z. Zinc Chelation with Hydroxamate in Histone Deacetylases Modulated by Water Access to the Linker Binding Channel. Journal of the American Chemical Society 133. [CrossRef]
- Baselious, F.; Hilscher, S.; Robaa, D.; Barinka, C.; Schutkowski, M.; Sippl, W. Comparative Structure-Based Virtual Screening Utilizing Optimized AlphaFold Model Identifies Selective HDAC11 Inhibitor. IJMS 2024, 25, 1358. [Google Scholar] [CrossRef]
- Baselious, F.; Robaa, D.; Sippl, W. Utilization of AlphaFold Models for Drug Discovery: Feasibility and Challenges. Histone Deacetylase 11 as a Case Study. Computers in Biology and Medicine 2023, 167, 107700. [Google Scholar] [CrossRef] [PubMed]
- Tretbar, M.; Schliehe-Diecks, J.; Von Bredow, L.; Tan, K.; Roatsch, M.; Tu, J.-W.; Kemkes, M.; Sönnichsen, M.; Schöler, A.; Borkhardt, A.; et al. Preferential HDAC6 Inhibitors Derived from HPOB Exhibit Synergistic Antileukemia Activity in Combination with Decitabine. European Journal of Medicinal Chemistry 2024, 272, 116447. [Google Scholar] [CrossRef] [PubMed]
- Ghazy, E.; Heimburg, T.; Lancelot, J.; Zeyen, P.; Schmidtkunz, K.; Truhn, A.; Darwish, S.; Simoben, C.V.; Shaik, T.B.; Erdmann, F.; et al. Synthesis, Structure-Activity Relationships, Cocrystallization and Cellular Characterization of Novel smHDAC8 Inhibitors for the Treatment of Schistosomiasis. European Journal of Medicinal Chemistry 2021, 225, 113745. [Google Scholar] [CrossRef] [PubMed]
- Ghazy, E.; Zeyen, P.; Herp, D.; Hügle, M.; Schmidtkunz, K.; Erdmann, F.; Robaa, D.; Schmidt, M.; Morales, E.R.; Romier, C.; et al. Design, Synthesis, and Biological Evaluation of Dual Targeting Inhibitors of Histone Deacetylase 6/8 and Bromodomain BRPF1. European Journal of Medicinal Chemistry 2020, 200, 112338. [Google Scholar] [CrossRef]
- Marek, M.; Shaik, T.B.; Heimburg, T.; Chakrabarti, A.; Lancelot, J.; Ramos-Morales, E.; Da Veiga, C.; Kalinin, D.; Melesina, J.; Robaa, D.; et al. Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants. J. Med. Chem. 2018, 61, 10000–10016. [Google Scholar] [CrossRef]
- Vögerl, K.; Ong, N.; Senger, J.; Herp, D.; Schmidtkunz, K.; Marek, M.; Müller, M.; Bartel, K.; Shaik, T.B.; Porter, N.J.; et al. Synthesis and Biological Investigation of Phenothiazine-Based Benzhydroxamic Acids as Selective Histone Deacetylase 6 Inhibitors. J. Med. Chem. 2019, 62, 1138–1166. [Google Scholar] [CrossRef]
- Uba, A.I.; Zengin, G. Phenolic Compounds as Histone Deacetylase Inhibitors: Binding Propensity and Interaction Insights from Molecular Docking and Dynamics Simulations. Amino Acids 2023, 55, 579–593. [Google Scholar] [CrossRef]
- Citarella, A.; Moi, D.; Pinzi, L.; Bonanni, D.; Rastelli, G. Hydroxamic Acid Derivatives: From Synthetic Strategies to Medicinal Chemistry Applications. ACS Omega 2021, 6, 21843–21849. [Google Scholar] [CrossRef] [PubMed]
- Riniker, S.; Landrum, G.A. Better Informed Distance Geometry: Using What We Know To Improve Conformation Generation. J. Chem. Inf. Model. 2015, 55, 2562–2574. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J Cheminform 2011, 3, 33. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J Comput Chem 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX : Tools for Structure Building and Analysis. Protein Science 2023, 32, e4792. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).