Submitted:
16 January 2025
Posted:
16 January 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Insight on TOR-Mediated Stress Responses via Metabolome Reprogramming
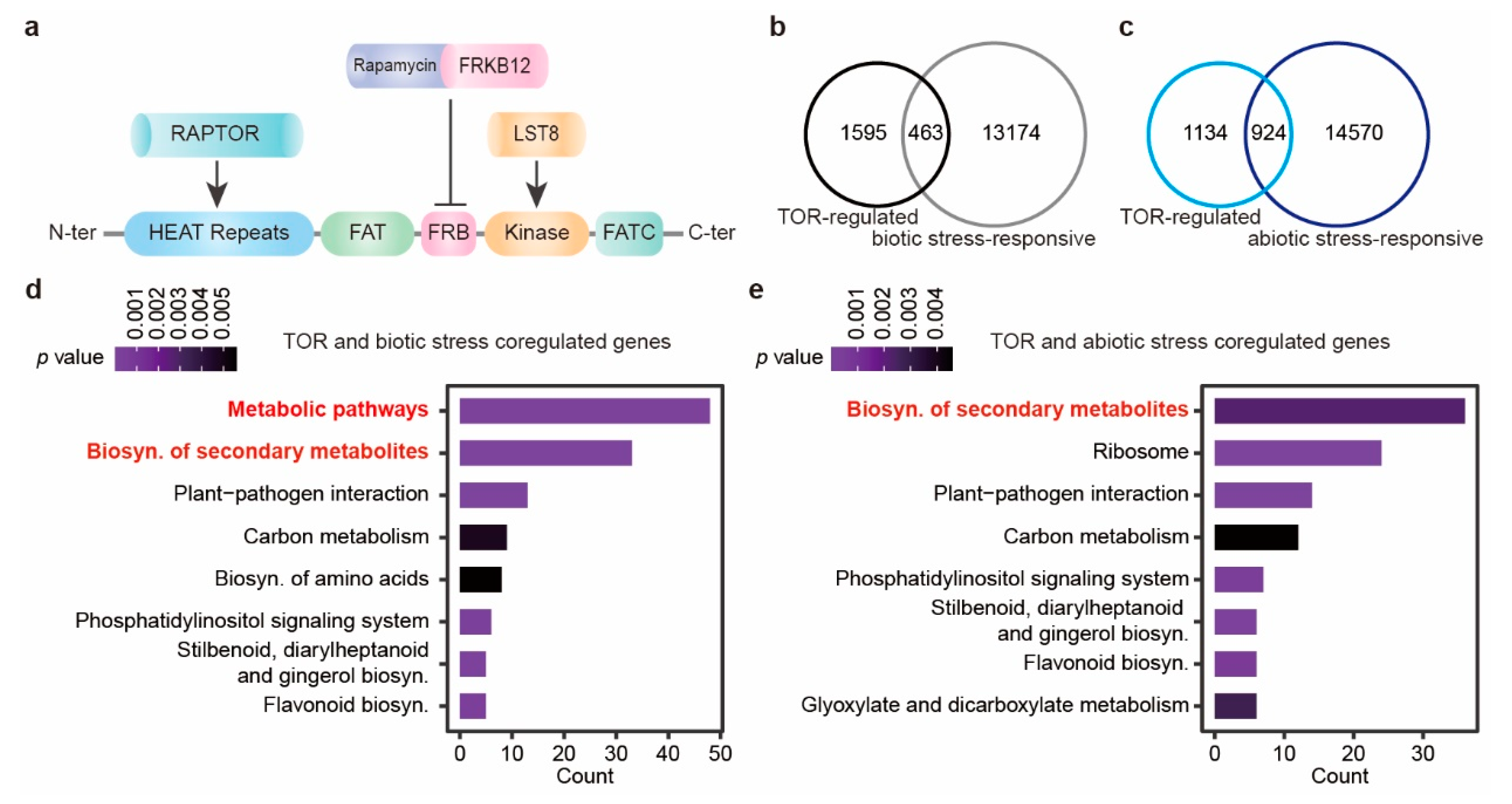
2.1. TOR Is Widely Involved in Plants Responses to Various Environmental Stresses
2.2. TOR-Regulated Plant Responses to Abiotic Stresses
2.3. TOR-Regulated Plant Responses to Biotic Stress
2.4. TOR-Modulated Metabolic Reprogramming Contributes to Plant Stress Responses
3. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, H., et al., Abiotic stress responses in plants. Nature Reviews Genetics, 2022. 23(2): p. 104-119. [CrossRef]
- Jones, J.D.G., B.J. Staskawicz, and J.L. Dangl, The plant immune system: From discovery to deployment. Cell, 2024. 187(9): p. 2095-2116. [CrossRef]
- Monson, R.K., et al., Coordinated resource allocation to plant growth-defense tradeoffs. New Phytol, 2022. 233(3): p. 1051-1066. [CrossRef]
- Liu, Y. and Y. Xiong, Plant target of rapamycin signaling network: Complexes, conservations, and specificities. Journal of Integrative Plant Biology, 2022. 64(2): p. 342-370. [CrossRef]
- Meng, Y., et al., TOR kinase, a GPS in the complex nutrient and hormonal signaling networks to guide plant growth and development. Journal of Experimental Botany, 2022. 73(20): p. 7041-7054. [CrossRef]
- Heitman, J., N.R. Movva, and M.N. Hall, Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science, 1991. 253(5022): p. 905-909. [CrossRef]
- Shi, F. and S. Collins, Regulation of mTOR signaling: Emerging role of cyclic nucleotide-dependent protein kinases and implications for cardiometabolic disease. Int J Mol Sci, 2023. 24(14): p. 11497.
- Emmerstorfer-Augustin, A. and J. Thorner, Regulation of TORC2 function and localization in yeast. Annu Rev Cell Dev Biol, 2023. 39: p. 363-389. [CrossRef]
- Wullschleger, S., R. Loewith, and M.N. Hall, TOR signaling in growth and metabolism. Cell, 2006. 124(3): p. 471-84. [CrossRef]
- Shi, L., Y. Wu, and J. Sheen, TOR signaling in plants: conservation and innovation. Development, 2018. 145(13). [CrossRef]
- Liu, G.Y. and D.M. Sabatini, mTOR at the nexus of nutrition, growth, ageing and disease. Nature Reviews Molecular Cell Biology, 2020. 21(4): p. 183-203. [CrossRef]
- Inoki, K., J. Kim, and K.L. Guan, AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol, 2012. 52: p. 381-400. [CrossRef]
- Burkart, G.M. and F. Brandizzi, A tour of TOR complex signaling in plants. Trends in Biochemical Sciences, 2021. 46(5): p. 417-428. [CrossRef]
- Saha, S., et al., mTORC1 and SGLT2 inhibitors-a therapeutic perspective for diabetic cardiomyopathy. Int J Mol Sci, 2023. 24(20): p. 15078.
- He, L., S. Cho, and J. Blenis, mTORC1, the maestro of cell metabolism and growth. Genes Dev, 2025. 39(1-2): p. 109-131. [CrossRef]
- Ragupathi, A., C. Kim, and E. Jacinto, The mTORC2 signaling network: Targets and cross-talks. Biochemical Journal, 2024. 481(2): p. 45-91. [CrossRef]
- Foltman, M. and A. Sanchez-Diaz, TOR complex 1: Orchestrating nutrient signaling and cell cycle progression. Int J Mol Sci, 2023. 24(21): p. 15745.
- Xiong, Y. and J. Sheen, Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. Journal of Biological Chemistry, 2012. 287(4): p. 2836-2842. [CrossRef]
- Xiong, Y., et al., Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature, 2013. 496(7444): p. 181-186. [CrossRef]
- Xiong, Y. and J. Sheen, The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiology, 2014. 164(2): p. 499-512. [CrossRef]
- Fu, L., P. Wang, and Y. Xiong, Target of rapamycin signaling in plant stress responses. Plant Physiology, 2020. 182(4): p. 1613-1623. [CrossRef]
- Xiong, Y. and J. Sheen, Moving beyond translation: glucose-TOR signaling in the transcriptional control of cell cycle. Cell Cycle, 2013. 12(13): p. 1989-90. [CrossRef]
- Xiong, Y., et al., Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature, 2013. 496(7444): p. 181-6. [CrossRef]
- Fu, L., et al., The TOR-EIN2 axis mediates nuclear signalling to modulate plant growth. Nature, 2021. 591(7849): p. 288-292. [CrossRef]
- Li, Z., et al., TOR balances plant growth and cold tolerance by orchestrating amino acid-derived metabolism in tomato. Hortic Res, 2024. 11(12): p. uhae253. [CrossRef]
- Hu, C., et al., Ethylene response factors 15 and 16 trigger jasmonate biosynthesis in tomato during herbivore resistance. Plant Physiol, 2021. 185(3): p. 1182-1197. [CrossRef]
- Naveed, Z.A. and G.S. Ali, Comparative transcriptome analysis between a resistant and a susceptible wild tomato accession in response to Phytophthora parasitica. Int J Mol Sci, 2018. 19(12). [CrossRef]
- Hu, Z., et al., High CO2- and pathogen-driven expression of the carbonic anhydrase βCA3 confers basal immunity in tomato. New Phytol, 2021. 229(5): p. 2827-2843. [CrossRef]
- Ikuyinminu, E., et al., Transcriptome, biochemical and phenotypic analysis of the effects of a precision engineered biostimulant for inducing salinity stress tolerance in tomato. Int J Mol Sci, 2023. 24(8). [CrossRef]
- Huang, Y., et al., HSFA1a modulates plant heat stress responses and alters the 3D chromatin organization of enhancer-promoter interactions. Nat Commun, 2023. 14(1): p. 469. [CrossRef]
- Liu, M., et al., Profiling of drought-responsive microRNA and mRNA in tomato using high-throughput sequencing. BMC Genomics, 2017. 18(1): p. 481. [CrossRef]
- Lin, R., et al., CALMODULIN6 negatively regulates cold tolerance by attenuating ICE1-dependent stress responses in tomato. Plant Physiol, 2023. 193(3): p. 2105-2121. [CrossRef]
- Ding, Y., Y. Shi, and S. Yang, Regulatory networks underlying plant responses and adaptation to cold stress. Annu Rev Genet, 2024. 58(1): p. 43-65. [CrossRef]
- Kan, Y., et al., The molecular basis of heat stress responses in plants. Molecular Plant, 2023. 16(10): p. 1612-1634. [CrossRef]
- Pereyra, C.M., et al., Target of rapamycin signaling is tightly and differently regulated in the plant response under distinct abiotic stresses. Planta, 2019. 251(1): p. 21. [CrossRef]
- Pacheco, J.M., et al., Cell surface receptor kinase FERONIA linked to nutrient sensor TORC signaling controls root hair growth at low temperature linked to low nitrate in Arabidopsis thaliana. New Phytol, 2023. 238(1): p. 169-185. [CrossRef]
- Wang, L., et al., The inhibition of protein translation mediated by AtGCN1 is essential for cold tolerance in Arabidopsis thaliana. Plant Cell Environ, 2017. 40(1): p. 56-68. [CrossRef]
- Dong, Y., et al., The Arabidopsis THADA homologue modulates TOR activity and cold acclimation. Plant Biol (Stuttg), 2019. 21 Suppl 1: p. 77-83. [CrossRef]
- Sharma, M., et al., Glucose-regulated HLP1 acts as a key molecule in governing thermomemory. Plant Physiol, 2019. 180(2): p. 1081-1100. [CrossRef]
- Sharma, M., et al., Arabidopsis target of rapamycin coordinates with transcriptional and epigenetic machinery to regulate thermotolerance. Front Plant Sci, 2021. 12: p. 741965. [CrossRef]
- Sharma, M., et al., A glucose-target of rapamycin signaling axis integrates environmental history of heat stress through maintenance of transcription-associated epigenetic memory in Arabidopsis. J Exp Bot, 2022. 73(20): p. 7083-7102. [CrossRef]
- Wang, C.F., et al., Plant salinity sensors: Current understanding and future directions. Front Plant Sci, 2022. 13: p. 859224. [CrossRef]
- Zhao, S., et al., Regulation of plant responses to salt stress. Int J Mol Sci, 2021. 22(9). [CrossRef]
- Deprost, D., et al., The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep, 2007. 8(9): p. 864-70. [CrossRef]
- Bakshi, A., et al., Target of Rapamycin (TOR) negatively regulates chlorophyll degradation and lipid peroxidation and controls responses under abiotic stress in Arabidopsis thaliana. Plant Stress, 2021. 2: p. 100020. [CrossRef]
- Kim, D., V.O. Ntui, and L. Xiong, Arabidopsis YAK1 regulates abscisic acid response and drought resistance. FEBS Lett, 2016. 590(14): p. 2201-2209. [CrossRef]
- Forzani, C., et al., Mutations of the AtYAK1 kinase suppress TOR deficiency in Arabidopsis. Cell Reports, 2019. 27(12): p. 3696-3708.e5. [CrossRef]
- Kavi Kishor, P.B., et al., Abscisic acid and its role in the modulation of plant growth, development, and yield stability. Trends Plant Sci, 2022. 27(12): p. 1283-1295. [CrossRef]
- Zhu, C., et al., How long should a kiss last between a kinase and its substrate? J Integr Plant Biol, 2022. 64(4): p. 789-791. [CrossRef]
- Wang, P., et al., Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol Cell, 2018. 69(1): p. 100-112.e6. [CrossRef]
- Jamsheer K, M., P. Awasthi, and A. Laxmi, The social network of target of rapamycin complex 1 in plants. Journal of Experimental Botany, 2022. 73(20): p. 7026-7040. [CrossRef]
- Mugume, Y., Z. Kazibwe, and D.C. Bassham, Target of rapamycin in control of autophagy: Puppet master and signal integrator. Int J Mol Sci, 2020. 21(21). [CrossRef]
- Soto-Burgos, J. and D.C. Bassham, SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS One, 2017. 12(8): p. e0182591. [CrossRef]
- Pu, Y., J. Soto-Burgos, and D.C. Bassham, Regulation of autophagy through SnRK1 and TOR signaling pathways. Plant Signal Behav, 2017. 12(12): p. e1395128. [CrossRef]
- Pu, Y., X. Luo, and D.C. Bassham, TOR-dependent and -independent pathways regulate autophagy in Arabidopsis thaliana. Front Plant Sci, 2017. 8: p. 1204. [CrossRef]
- Liu, Y. and D.C. Bassham, Autophagy: Pathways for self-eating in plant cells. Annu Rev Plant Biol, 2012. 63: p. 215-37. [CrossRef]
- Van Leene, J., et al., Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat Plants, 2019. 5(3): p. 316-327. [CrossRef]
- Jhu, M.Y. and N.R. Sinha, Parasitic plants: An overview of mechanisms by which plants perceive and respond to parasites. Annu Rev Plant Biol, 2022. 73: p. 433-455. [CrossRef]
- Shelake, R.M., et al., Heat stress and plant-biotic interactions: Advances and perspectives. Plants (Basel), 2024. 13(15): p. 2022.
- Zhang, Y., et al., SlMYC2 interacted with the SlTOR promoter and mediated JA signaling to regulate growth and fruit quality in tomato. Front Plant Sci, 2022. 13: p. 1013445. [CrossRef]
- Farooq, M.A., et al., Jasmonic acid mediates Ca2+ dependent signal transduction and plant immunity. Plant Sci, 2024. 348: p. 112239. [CrossRef]
- Meteignier, L.V., et al., Translatome analysis of an NB-LRR immune response identifies important contributors to plant immunity in Arabidopsis. J Exp Bot, 2017. 68(9): p. 2333-2344. [CrossRef]
- Popa, C., et al., The effector AWR5 from the plant pathogen Ralstonia solanacearum is an inhibitor of the TOR signalling pathway. Sci Rep, 2016. 6: p. 27058. [CrossRef]
- Marash, I., et al., TOR inhibition primes immunity and pathogen resistance in tomato in a salicylic acid-dependent manner. Mol Plant Pathol, 2022. 23(7): p. 1035-1047. [CrossRef]
- Marash, I., et al., TOR coordinates cytokinin and gibberellin signals mediating development and defense. Plant Cell Environ, 2024. 47(2): p. 629-650. [CrossRef]
- De Vleesschauwer, D., et al., Target of rapamycin signaling orchestrates growth-defense trade-offs in plants. New Phytol, 2018. 217(1): p. 305-319. [CrossRef]
- Zhang, H., et al., Different viral effectors suppress hormone-mediated antiviral immunity of rice coordinated by OsNPR1. Nat Commun, 2023. 14(1): p. 3011. [CrossRef]
- Aznar, N.R., et al., TOR signaling downregulation increases resistance to the cereal killer Fusarium graminearum. Plant Signal Behav, 2018. 13(2): p. e1414120. [CrossRef]
- Schepetilnikov, M., et al., Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. Embo j, 2011. 30(7): p. 1343-56. [CrossRef]
- Ouibrahim, L., et al., Potyviruses differ in their requirement for TOR signalling. J Gen Virol, 2015. 96(9): p. 2898-2903. [CrossRef]
- Szwed, A., E. Kim, and E. Jacinto, Regulation and metabolic functions of mTORC1 and mTORC2. Physiol Rev, 2021. 101(3): p. 1371-1426. [CrossRef]
- Li, L., et al., Target of rapamycin controls hyphal growth and pathogenicity through FoTIP4 in Fusarium oxysporum. Mol Plant Pathol, 2021. 22(10): p. 1239-1255. [CrossRef]
- Calderan-Rodrigues, M.J., et al., Proteogenic dipeptides are characterized by diel fluctuations and target of rapamycin complex-signaling dependency in the model plant Arabidopsis thaliana. Front Plant Sci, 2021. 12: p. 758933. [CrossRef]
- da Silva, V.C.H., et al., Shedding light on the dynamic role of the "Target of Rapamycin" kinase in the fast-growing C4 species Setaria viridis, a suitable model for biomass crops. Front Plant Sci, 2021. 12: p. 637508. [CrossRef]
- Busche, M., et al., TOR coordinates nucleotide availability with ribosome biogenesis in plants. Plant Cell, 2021. 33(5): p. 1615-1632. [CrossRef]
- Mubeen, U., P. Giavalisco, and C. Caldana, TOR inhibition interrupts the metabolic homeostasis by shifting the carbon-nitrogen balance in Chlamydomonas reinhardtii. Plant Signal Behav, 2019. 14(11): p. 1670595. [CrossRef]
- Scarpin, M.R., S. Leiboff, and J.O. Brunkard, Parallel global profiling of plant TOR dynamics reveals a conserved role for LARP1 in translation. Elife, 2020. 9. [CrossRef]
- Roustan, V. and W. Weckwerth, Quantitative phosphoproteomic and system-level analysis of TOR inhibition unravel distinct organellar acclimation in Chlamydomonas reinhardtii. Front Plant Sci, 2018. 9: p. 1590. [CrossRef]
- Han, C., et al., TOR promotes guard cell starch degradation by regulating the activity of β-AMYLASE1 in Arabidopsis. Plant Cell, 2022. 34(3): p. 1038-1053. [CrossRef]
- Moreau, M., et al., Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell, 2012. 24(2): p. 463-81. [CrossRef]
- Li, D., et al., Target of rapamycin (TOR) regulates the response to low nitrogen stress via autophagy and hormone pathways in Malus hupehensis. Hortic Res, 2022. 9: p. uhac143. [CrossRef]
- Ren, M., et al., Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell, 2012. 24(12): p. 4850-74. [CrossRef]
- Li, L., et al., Functional characterization of target of rapamycin signaling in Verticillium dahliae. Front Microbiol, 2019. 10: p. 501. [CrossRef]
- Song, L., et al., The RALF1-FERONIA complex interacts with and activates TOR signaling in response to low nutrients. Mol Plant, 2022. 15(7): p. 1120-1136. [CrossRef]
- Mubeen, U., et al., Target of rapamycin inhibition in Chlamydomonas reinhardtii triggers de novo amino acid synthesis by enhancing nitrogen assimilation. Plant Cell, 2018. 30(10): p. 2240-2254. [CrossRef]
- Caldana, C., et al., Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J, 2013. 73(6): p. 897-909. [CrossRef]
- Forzani, C., et al., Mutations of the AtYAK1 kinase suppress TOR deficiency in Arabidopsis. Cell Rep, 2019. 27(12): p. 3696-3708.e5. [CrossRef]
- O'Leary, B.M., et al., Metabolite regulatory interactions control plant respiratory metabolism via target of rapamycin (TOR) kinase activation. Plant Cell, 2020. 32(3): p. 666-682. [CrossRef]
- Artins, A., et al., Sensing and regulation of C and N metabolism-novel features and mechanisms of the TOR and SnRK1 signaling pathways. Plant J, 2024. 118(5): p. 1268-1280. [CrossRef]
- Ding, Y. and S. Yang, Surviving and thriving: How plants perceive and respond to temperature stress. Dev Cell, 2022. 57(8): p. 947-958. [CrossRef]
- Jancewicz, A.L., N.M. Gibbs, and P.H. Masson, Cadaverine's functional role in plant development and environmental response. Front Plant Sci, 2016. 7: p. 870. [CrossRef]
- Ozmen, S., S. Tabur, and S. Oney-Birol, Alleviation role of exogenous cadaverine on cell cycle, endogenous polyamines amounts and biochemical enzyme changes in barley seedlings under drought stress. Scientific Reports, 2023. 13(1): p. 17488. [CrossRef]
- You, J., et al., Transcriptomic and metabolomic profiling of drought-tolerant and susceptible sesame genotypes in response to drought stress. BMC Plant Biol, 2019. 19(1): p. 267. [CrossRef]
- Barqawi, A.A. and A.A. Abulfaraj, Salt stress-related mechanisms in leaves of the wild barley Hordeum spontaneum generated from RNA-Seq datasets. Life (Basel), 2023. 13(7). [CrossRef]
- Brocker, C., et al., Aldehyde dehydrogenase 7A1 (ALDH7A1) attenuates reactive aldehyde and oxidative stress induced cytotoxicity. Chem Biol Interact, 2011. 191(1-3): p. 269-77. [CrossRef]
- Masclaux-Daubresse, C., et al., Stitching together the multiple dimensions of autophagy using metabolomics and transcriptomics reveals impacts on metabolism, development, and plant responses to the environment in Arabidopsis. Plant Cell, 2014. 26(5): p. 1857-1877. [CrossRef]
- Huh, S.U., et al., ATAF2, a NAC transcription factor, binds to the promoter and regulates NIT2 gene expression involved in auxin biosynthesis. Mol Cells, 2012. 34(3): p. 305-13. [CrossRef]
- Kwon, Y.S., et al., Proteomic analyses of the interaction between the plant-growth promoting rhizobacterium Paenibacillus polymyxa E681 and Arabidopsis thaliana. Proteomics, 2016. 16(1): p. 122-35. [CrossRef]
- Luo, K., et al., Indole-3-acetic acid in Fusarium graminearum: Identification of biosynthetic pathways and characterization of physiological effects. Fungal Biol, 2016. 120(9): p. 1135-45. [CrossRef]
- Alsherif, E.A., et al., How carbon nanoparticles, arbuscular mycorrhiza, and compost mitigate drought stress in maize plant: A growth and biochemical study. Plants (Basel), 2022. 11(23). [CrossRef]
- Hijaz, F. and N. Killiny, Exogenous GABA is quickly metabolized to succinic acid and fed into the plant TCA cycle. Plant Signal Behav, 2019. 14(3): p. e1573096. [CrossRef]
- Zhou, H., et al., Potassium indole-3-butyric acid affects rice's adaptability to salt stress by regulating carbon metabolism, transcription factor genes expression, and biosynthesis of secondary metabolites. Front Plant Sci, 2024. 15: p. 1416936. [CrossRef]
- Yang, L., et al., Hydroxycoumarins: New, effective plant-derived compounds reduce Ralstonia pseudosolanacearum populations and control tobacco bacterial wilt. Microbiol Res, 2018. 215: p. 15-21. [CrossRef]
- Wang, Y., et al., 5-ALA, DTA-6, and nitrogen mitigate NaCl stress by promoting photosynthesis and carbon metabolism in rice seedlings. Metabolites, 2024. 14(3): p. 142.
- Rhaman, M.S., et al., 5-aminolevulinic acid-mediated plant adaptive responses to abiotic stress. Plant Cell Rep, 2021. 40(8): p. 1451-1469. [CrossRef]
- Tan, S., et al., Advances in 5-aminolevulinic acid priming to enhance plant tolerance to abiotic stress. Int J Mol Sci, 2022. 23(2). [CrossRef]
- El-Shora, H.M., et al., Alleviation of lead stress on sage plant by 5-aminolevulinic acid (ALA). Plants (Basel), 2021. 10(9). [CrossRef]
- Yang, Y., et al., Copper stress in grapevine: Consequences, responses, and a novel mitigation strategy using 5-aminolevulinic acid. Environ Pollut, 2022. 307: p. 119561. [CrossRef]
- Yang, Y., et al., Drought stress in 'Shine Muscat' grapevine: Consequences and a novel mitigation strategy-5-aminolevulinic acid. Front Plant Sci, 2023. 14: p. 1129114. [CrossRef]
- Helaly, M.N., et al., 5-aminolevulinic acid and 24-epibrassinolide improve the drought stress resilience and productivity of banana plants. Plants (Basel), 2022. 11(6): p. 743.
- Yu, J., et al., Metabolic pathways involved in carbon dioxide enhanced heat tolerance in bermudagrass. Front Plant Sci, 2017. 8: p. 1506. [CrossRef]
- Lei, S., et al., Metabolic regulation of 5-oxoproline for enhanced heat tolerance in perennial ryegrass. Stress Biol, 2024. 4(1): p. 46. [CrossRef]
- Wang, Y., et al., Metabolite profiling in two contrasting Tibetan hulless barley cultivars revealed the core salt-responsive metabolome and key salt-tolerance biomarkers. AoB Plants, 2019. 11(2): p. plz021. [CrossRef]
- Bruňáková, K., et al., Does phenotyping of Hypericum secondary metabolism reveal a tolerance to biotic/abiotic stressors? Front Plant Sci, 2022. 13: p. 1042375. [CrossRef]
- Wu, Z., et al., Study of dandelion (Taraxacum mongolicum Hand.-Mazz.) salt response and caffeic acid metabolism under saline stress by transcriptome analysis. Genes (Basel), 2024. 15(2). [CrossRef]
- Ramzan, M., et al., Potential of kaempferol and caffeic acid to mitigate salinity stress and improving potato growth. Sci Rep, 2024. 14(1): p. 21657. [CrossRef]
- Li, J., et al., Strigolactone enhances tea plant adaptation to drought and Phyllosticta theicola petch by regulating caffeine content via CsbHLH80. Plant Physiol Biochem, 2024. 216: p. 109161. [CrossRef]
- Yoo, Y., et al., Caffeine produced in rice plants provides tolerance to water-deficit stress. Antioxidants (Basel), 2023. 12(11). [CrossRef]
- Zhang, N., et al., Constitutive camalexin production and environmental stress response variation in Arabidopsis populations from the Iberian Peninsula. Plant Sci, 2014. 225: p. 77-85. [CrossRef]
- Xiao, P., et al., Transcriptome and metabolome atlas reveals contributions of sphingosine and chlorogenic acid to cold tolerance in Citrus. Plant Physiol, 2024. 196(1): p. 634-650. [CrossRef]
- Piasecka, A., et al., Phenolic metabolites from barley in contribution to phenome in soil moisture deficit. Int J Mol Sci, 2020. 21(17). [CrossRef]
- Dias, M.C., et al., Phenolic and lipophilic metabolite adjustments in Olea europaea (olive) trees during drought stress and recovery. Phytochemistry, 2021. 185: p. 112695. [CrossRef]
- Lacrampe, N., et al., Nitrogen-mediated metabolic patterns of susceptibility to Botrytis cinerea infection in tomato (Solanum lycopersicum) stems. Planta, 2023. 257(2): p. 41. [CrossRef]
- Silambarasan, S., et al., Co-application of citric acid and Nocardiopsis sp. strain RA07 enhances phytoremediation potentiality of Sorghum bicolor L. Environ Sci Pollut Res Int, 2023. 30(36): p. 86244-86254. [CrossRef]
- Menhas, S., et al., Citric acid-driven cadmium uptake and growth promotion mechanisms in Brassica napus. Chemosphere, 2024. 368: p. 143716. [CrossRef]
- Cay, S., Assessment of tea saponin and citric acid-assisted phytoextraction of Pb-contaminated soil by Salvia virgata Jacq. Environ Sci Pollut Res Int, 2023. 30(17): p. 49771-49778. [CrossRef]
- Kaya, C., et al., Citric acid and hydrogen sulfide cooperate to mitigate chromium stress in tomato plants by modulating the ascorbate-glutathione cycle, chromium sequestration, and subcellular allocation of chromium. Environ Pollut, 2023. 335: p. 122292. [CrossRef]
- Tahjib-Ul-Arif, M., et al., Citric acid-mediated abiotic stress tolerance in plants. Int J Mol Sci, 2021. 22(13). [CrossRef]
- Święcicka, M., et al., Changes in benzoxazinoid contents and the expression of the associated genes in rye (Secale cereale L.) due to brown rust and the inoculation procedure. PLoS One, 2020. 15(5): p. e0233807. [CrossRef]
- Zhang, X., et al., Mechanisms of resistance to spot blotch in Yunnan iron shell wheat based on metabolome and transcriptomics. Int J Mol Sci, 2022. 23(9). [CrossRef]
- Rao, M.J., et al., LC-MS/MS-based metabolomics approach identified novel antioxidant flavonoids associated with drought tolerance in citrus species. Front Plant Sci, 2023. 14: p. 1150854. [CrossRef]
- Kim, S.E., et al., Overexpression of 4-hydroxyphenylpyruvate dioxygenase (IbHPPD) increases abiotic stress tolerance in transgenic sweetpotato plants. Plant Physiol Biochem, 2021. 167: p. 420-429. [CrossRef]
- Mata-Pérez, C., et al., Protein tyrosine nitration during development and abiotic stress response in plants. Front Plant Sci, 2016. 7: p. 1699. [CrossRef]
- Mishra, S., A. Sharma, and A.K. Srivastava, Ascorbic acid: a metabolite switch for designing stress-smart crops. Crit Rev Biotechnol, 2024. 44(7): p. 1350-1366. [CrossRef]
- Biswas, S., A.K. Biswas, and B. De, Influence of sodium chloride on growth and metabolic reprogramming in nonprimed and haloprimed seedlings of blackgram (Vigna mungo L.). Protoplasma, 2020. 257(6): p. 1559-1583. [CrossRef]
- Li, X., et al., Integrative physiological, metabolomic, and transcriptomic analysis reveals the drought responses of two apple rootstock cultivars. BMC Plant Biol, 2024. 24(1): p. 219. [CrossRef]
- Zeier, J., New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ, 2013. 36(12): p. 2085-103. [CrossRef]
- Zou, Y.N., et al., Metabolomics reveals arbuscular mycorrhizal fungi-mediated tolerance of walnut to soil drought. BMC Plant Biol, 2023. 23(1): p. 118. [CrossRef]
- Wang, Y., et al., Transcriptomics and metabolomics revealed that phosphate improves the cold tolerance of alfalfa. Front Plant Sci, 2023. 14: p. 1100601. [CrossRef]
- Castro-Vázquez, L., et al., Pressurized extraction as an opportunity to recover antioxidants from orange peels: Heat treatment and nanoemulsion design for modulating oxidative stress. Molecules, 2021. 26(19). [CrossRef]
- Lwalaba, J.L.W., et al., Transcriptome analysis reveals the tolerant mechanisms to cobalt and copper in barley. Ecotoxicol Environ Saf, 2021. 209: p. 111761. [CrossRef]
- Jin, S., et al., Accumulation of hydroxycinnamic acid amides in winter wheat under snow. Biosci Biotechnol Biochem, 2003. 67(6): p. 1245-9. [CrossRef]
- Neamțu, A.A., et al., A comprehensive view on the impact of chlorogenic acids on colorectal cancer. Curr Issues Mol Biol, 2024. 46(7): p. 6783-6804. [CrossRef]
- Huo, X., et al., Effect of S-Allyl-L-Cysteine on nitric oxide and Cadmium processes in Rice (Oryza sativa L. sp. Zhongzao35) seedlings. Toxics, 2024. 12(11). [CrossRef]
- Cheng, L.L., et al., Mechanism of S-allyl-L-cysteine alleviating Cadmium stress in seedling roots and buds of rice seedlings. Huan Jing Ke Xue, 2021. 42(6): p. 3037-3045. [CrossRef]
- Owusu, A.G., et al., Transcriptomic and metabolomic analyses reveal the potential mechanism of waterlogging resistance in cotton (Gossypium hirsutum L.). Front Plant Sci, 2023. 14: p. 1088537. [CrossRef]
- Zhuang, Q., et al., Joint transcriptomic and metabolomic analysis reveals the mechanism of low-temperature tolerance in Hosta ventricosa. PLoS One, 2021. 16(11): p. e0259455. [CrossRef]
- Zhou, C., et al., Novel finding on how melatonin and nanoselenium alleviate 2,4-D butylate stress in wheat plants. J Agric Food Chem, 2023. 71(35): p. 12943-12957. [CrossRef]
- Dong, X., et al., Multiomics analyses reveal MsC3H29 positively regulates flavonoid biosynthesis to improve drought resistance of autotetraploid cultivated alfalfa (Medicago sativa L.). J Agric Food Chem, 2024. 72(25): p. 14448-14465. [CrossRef]
- Kasthuri, T., et al., Proteomic profiling spotlights the molecular targets and the impact of the natural antivirulent umbelliferone on stress response, virulence factors, and the quorum sensing network of Pseudomonas aeruginosa. Front Cell Infect Microbiol, 2022. 12: p. 998540. [CrossRef]
- Beesley, A., et al., Engineered coumarin accumulation reduces mycotoxin-induced oxidative stress and disease susceptibility. Plant Biotechnol J, 2023. 21(12): p. 2490-2506. [CrossRef]
- Zhou, Y., et al., Transcriptome sequencing and metabolome analysis reveal the molecular mechanism of Salvia miltiorrhiza in response to drought stress. BMC Plant Biol, 2024. 24(1): p. 446. [CrossRef]
| Compounds | Class | Up- or Down-regulated in tor-es | Function | References |
|---|---|---|---|---|
| 1,5-Diaminopentane | Phenolamides | up | Drought, oxidative stress, heavy metal stress | [91,92] |
| 2-Aminoadipic acid (L-Homoglutamic acid) | Amino acid and derivatives | up | Oxidative stress, drought, salt | [93,94,95,96] |
| 3-Indoleacetonitrile | Indole derivatives | down | Pathogen attack | [97,98,99] |
| 3-Methylmalic acid | Organic acids and derivatives | up | Salt, drought, biotic stress | [100,101,102] |
| 4-Hydroxycoumarin | Phenylpropanoids | down | Pathogen attack | [103] |
| 5-Aminolevulinate | Organic acids and derivatives | up | Drought, salt, heavy metal | [104,105,106,107,108,109,110] |
| 5-Oxoproline | Amino acid and derivatives | up | Heat stress | [111,112] |
| 6-Aminocaproic acid | Organic acids and derivatives | up | Salt stress | [113] |
| Amentoflavone | Flavone | up | Temperature, light, drought, biotic stress | [114] |
| Caffeic acid | Phenylpropanoids | up | Salt stress | [115,116] |
| Caffeine | Alkaloids | down | Drought, biotic stress | [117,118] |
| Camalexin | Alkaloids | up | Pathogen attack | [119] |
| Chlorogenic acid (3-O-Caffeoylquinic acid) | Organic acids and derivatives | down | Cold stress | [120] |
| Chrysoeriol | Flavone | up | Oxidative stress | [121,122] |
| Citramalate | Organic acids and derivatives | up | Pathogen attack | [123] |
| Citric acid monohydrate | Organic acids and derivatives | up | Heavy metal | [124,125,126,127,128] |
| Citric acid | Organic acids and derivatives | up | Heavy metal | [124,125,126,127,128] |
| DIMBOA glucoside | Others | up | Biotic stress | [129,130] |
| Genistein 7-O-Glucoside (Genistin) | Isoflavone | up | Drought | [131] |
| Homogentisic acid | Organic acids and derivatives | up | Abiotic stress, ABA signalling | [132] |
| L-(-)-Tyrosine | Amino acid and derivatives | up | Abiotic stress | [133] |
| L-Ascorbate | Vitamins and derivatives | up | Abiotic stress | [134] |
| L-Homoserine | Amino acid and derivatives | up | Drought, salt stress | [135,136] |
| L-Pipecolic acid | Amino acid and derivatives | up | Biotic stress | [137] |
| N-Acetyl-L-phenylalanine | Amino acid and derivatives | up | Cold, drought | [138,139] |
| Narirutin | Flavone | up | Heavy metal stress, oxidative stress | [140,141] |
| N-p-Coumaroyl agmatine | Phenolamides | up | Biotic stress | [142] |
| Quinic acid | Organic acids and derivatives | up | Abiotic stress | [143] |
| S-Allyl-L-cysteine | Amino acid and derivatives | up | Heavy metal stress | [144,145] |
| Sinapyl alcohol | Phenylpropanoids | up | Flooding stress, cold stress | [146,147] |
| Tricin | Flavone | down | Cold, drought, salt stress | [148,149] |
| Umbelliferone | Phenylpropanoids | down | Pathogen attack | [150,151] |
| Xanthohumol | Flavanone | up | Drought stress | [152] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





