Submitted:
10 February 2025
Posted:
11 February 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Determination of msp2/p44 Repertoires
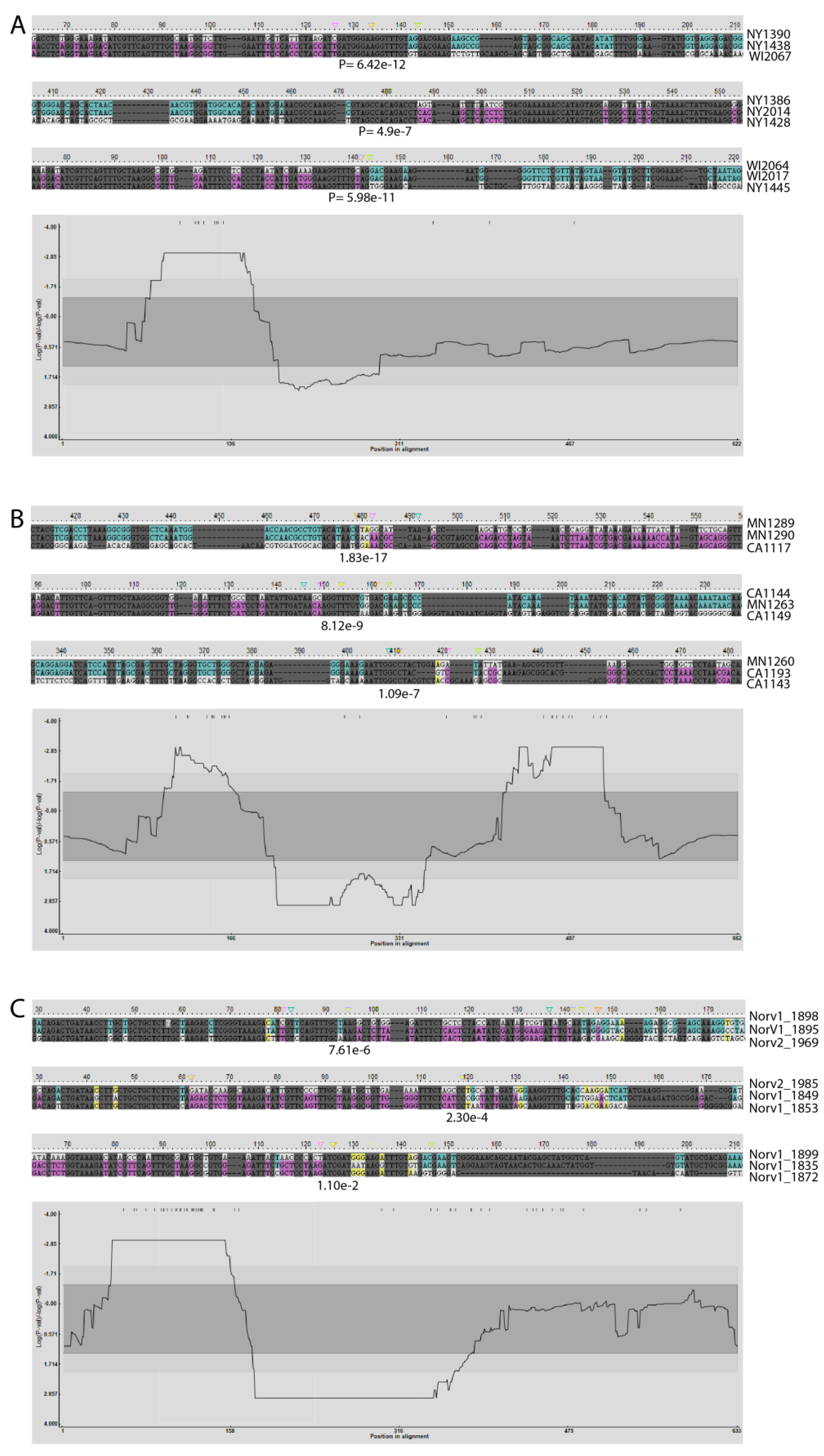
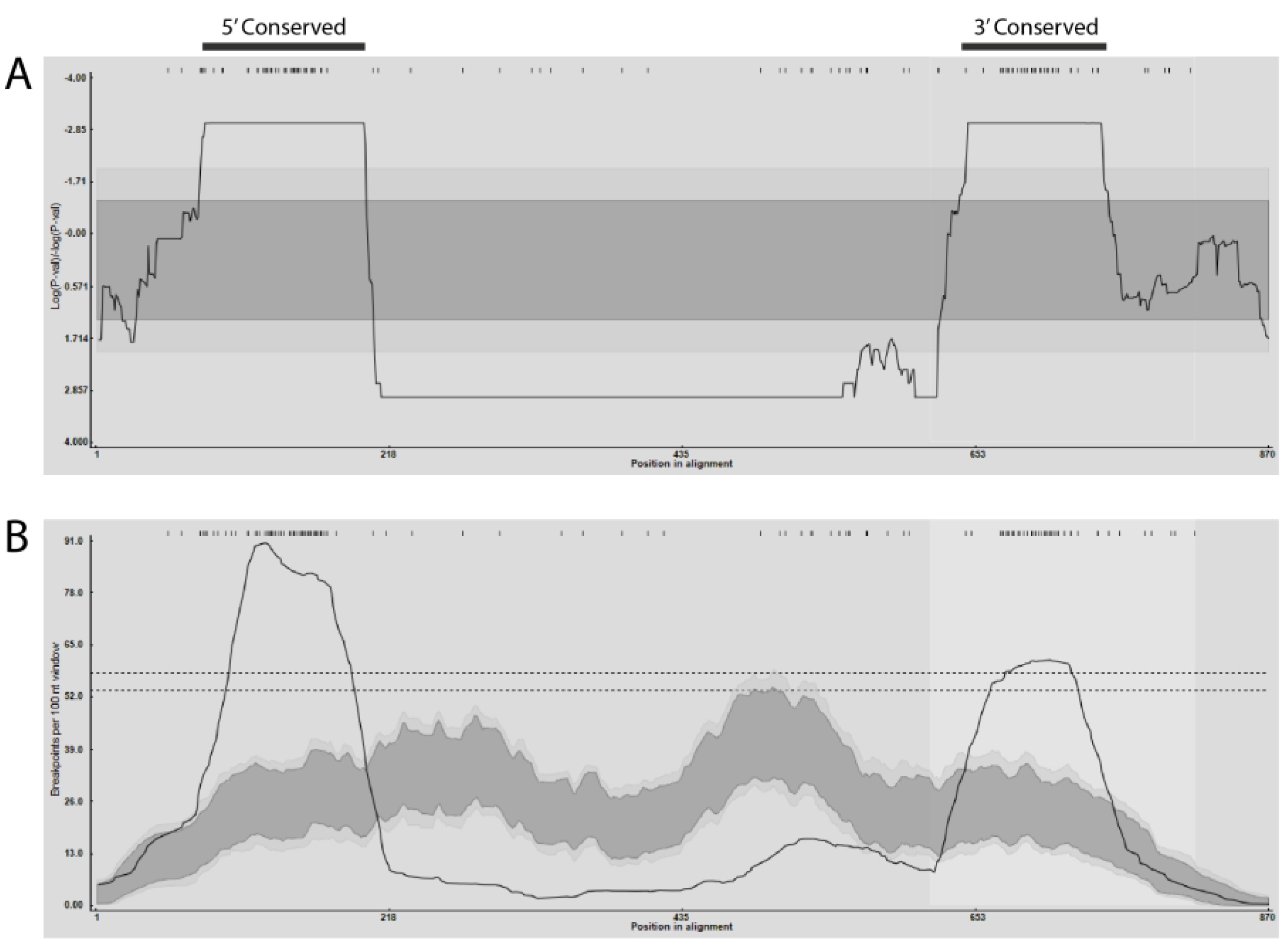
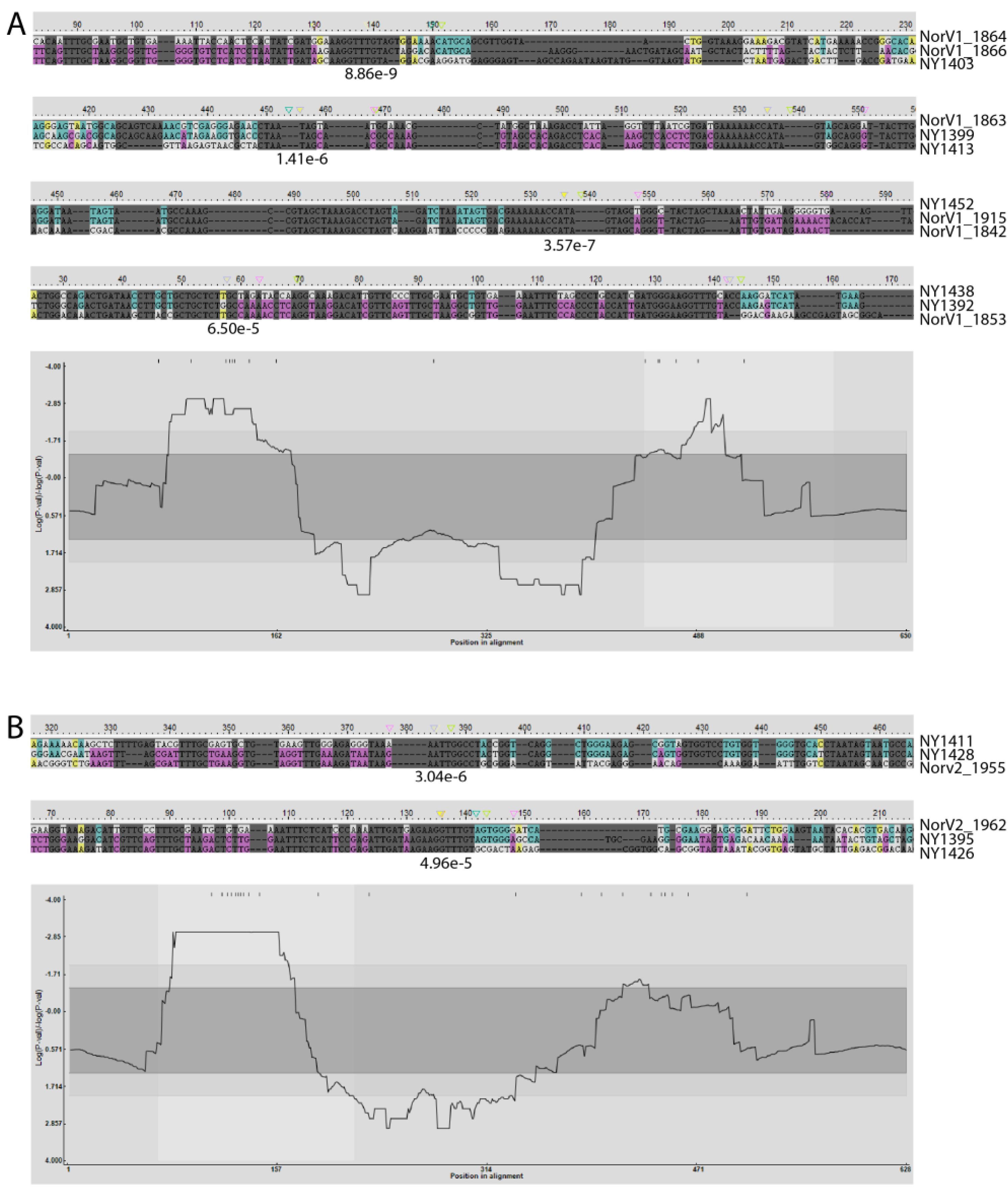
2.2. Detection of Recombination
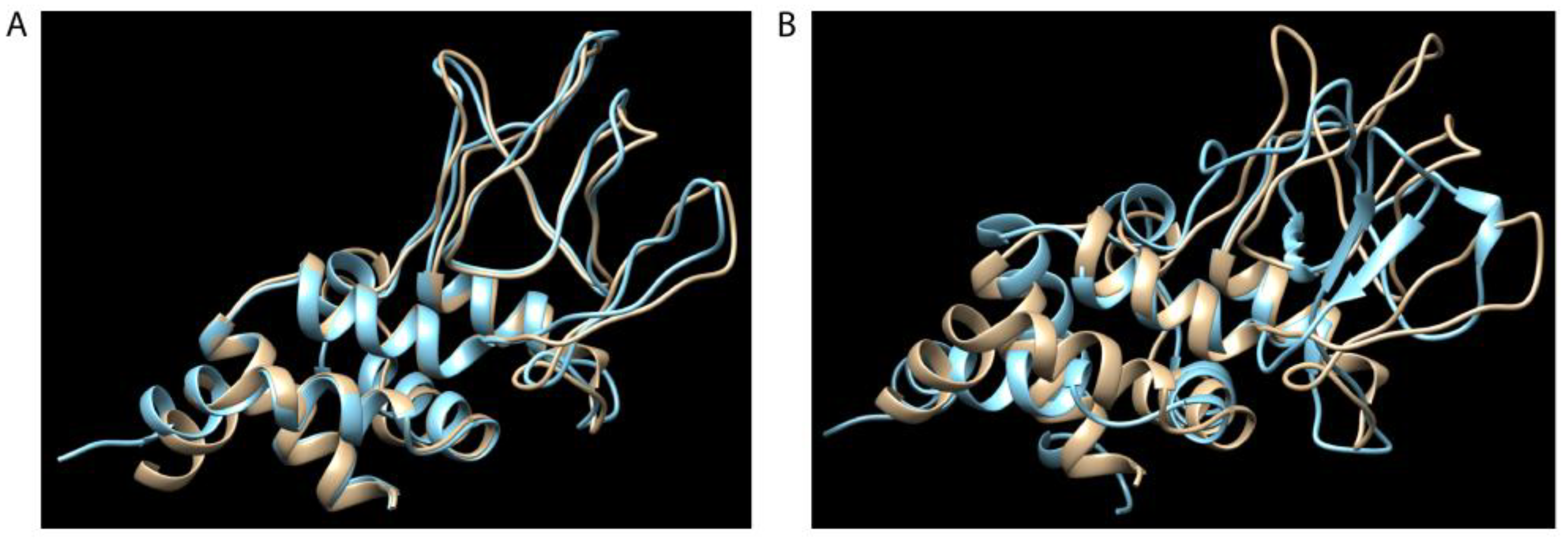
2.3. Polypeptide Structural Comparisons
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CVR | Central variable region of msp2/p44 genes |
| MSP2 | Major surface protein 2 |
References
- Stuen, S. Anaplasma phagocytophilum - the most widespread tick-borne infection in animals in Europe. Vet Res Commun. 2007, 31 (Suppl. 1), 79–84. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.C.; Barbet, A.F. Persistent Infections and immunity in ruminants to arthropod-borne bacteria in the family Anaplasmataceae. Annu Rev Anim Biosci. 2016, 4, 177–197. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kikuchi, T.; Rikihisa, Y. Two monoclonal antibodies with defined epitopes of P44 major surface proteins neutralize Anaplasma phagocytophilum by distinct mechanisms. Infect Immun. 2006, 74, 1873–1882. [Google Scholar] [CrossRef]
- Wang, X.; Rikihisa, Y.; Lai, T.H.; Kumagai, Y.; Zhi, N.; Reed, S.M. Rapid sequential changeover of expressed p44 genes during the acute phase of Anaplasma phagocytophilum infection in horses. Infect Immun. 2004, 72, 6852–6859. [Google Scholar] [CrossRef]
- Granquist, E.G.; Stuen, S.; Crosby, L.; Lundgren, A.M.; Alleman, A.R.; Barbet, A.F. Variant-specific and diminishing immune responses towards the highly variable MSP2(P44) outer membrane protein of Anaplasma phagocytophilum during persistent infection in lambs. Vet Immunol Immunopathol. 2010, 133, 117–124. [Google Scholar] [CrossRef]
- Granquist, E.G.; Stuen, S.; Lundgren, A.M.; Bråten, M.; Barbet, A.F. Outer membrane protein sequence variation in lambs experimentally infected with Anaplasma phagocytophilum. Infect Immun. 2008, 76, 120–126. [Google Scholar] [CrossRef]
- Barbet, A.F.; Meeus, P.F.; Bélanger, M.; Bowie, M.V.; Yi, J.; Lundgren, A.M.; et al. Expression of multiple outer membrane protein sequence variants from a single genomic locus of Anaplasma phagocytophilum. Infect Immun. 2003, 71, 1706–1718. [Google Scholar] [CrossRef]
- Lin, Q.; Rikihisa, Y. Establishment of cloned Anaplasma phagocytophilum and analysis of p44 gene conversion within an infected horse and infected SCID mice. Infect Immun. 2005, 73, 5106–5114. [Google Scholar] [CrossRef]
- Lin, Q.; Zhang, C.; Rikihisa, Y. Analysis of involvement of the RecF pathway in p44 recombination in Anaplasma phagocytophilum and in Escherichia coli by using a plasmid carrying the p44 expression and p44 donor loci. Infect Immun. 2006, 74, 2052–2062. [Google Scholar] [CrossRef]
- Lin, Q.; Ohashi, N.; Horowitz, H.W.; Aguero-Rosenfeld, M.E.; Raffalli, J.; Wormser, G.P.; et al. Analysis of sequences and loci of p44 homologs expressed by Anaplasma phagocytophila in acutely infected patients. J Clin Microbiol. 2002, 40, 2981–2988. [Google Scholar] [CrossRef]
- Dunning Hotopp, J.C.; Lin, M.; Madupu, R.; Crabtree, J.; Angiuoli, S.V.; Eisen, J.A.; et al. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2006, 2, e21. [Google Scholar]
- Brayton, K.A.; Kappmeyer, L.S.; Herndon, D.R.; Dark, M.J.; Tibbals, D.L.; Palmer, G.H.; et al. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci USA. 2005, 102, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Palmer, G.H.; Bankhead, T.; Seifert, H.S. Antigenic variation in bacterial pathogens. Microbiol Spectrum. 2016, 4. [Google Scholar] [CrossRef]
- Barbet, A.F.; Lundgren, A.M.; Yi, J.; Rurangirwa, F.R.; Palmer, G.H. Antigenic variation of Anaplasma marginale by expression of MSP2 mosaics. Infect Immun. 2000, 68, 6133–6138. [Google Scholar] [CrossRef]
- Futse, J.E.; Brayton, K.A.; Knowles, D.P.; Palmer, G.H. Structural basis for segmental gene conversion in generation of Anaplasma marginale outer membrane protein variants. Mol Microbiol. 2005, 57, 212–221. [Google Scholar] [CrossRef]
- Rejmanek, D.; Foley, P.; Barbet, A.F.; Foley, J. Evolution of antigen variation in the tick-borne pathogen Anaplasma phagocytophilum. Mol Biol Evol. 2012, 29, 391–400. [Google Scholar] [CrossRef]
- Lin, Q.; Rikihisa, Y.; Ohashi, N.; Zhi, N. Mechanisms of variable p44 expression by Anaplasma phagocytophilum. Infect Immun. 2003, 71, 5650–5661. [Google Scholar] [CrossRef]
- Crosby, F.L.; Eskeland, S.; Bø-Granquist, E.G.; Munderloh, U.G.; Price, L.D.; Al-Khedery, B.; et al. Comparative whole genome analysis of an Anaplasma phagocytophilum strain isolated from Norwegian sheep. Pathogens. 2022, 11, 601. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; et al. RDP5, a computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef]
- Barbet, A.F.; Al-Khedery, B.; Stuen, S.; Granquist, E.G.; Felsheim, R.F.; Munderloh, U.G. An emerging tick-borne disease of humans is caused by a subset of strains with conserved genome structure. Pathogens. 2013, 2, 544–555. [Google Scholar] [CrossRef]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Research. 2004, 32 Suppl 2, W526–W31. [Google Scholar] [CrossRef]
- Park, J.; Choi, K.S.; Dumler, J.S. Major surface protein 2 of Anaplasma phagocytophilum facilitates adherence to granulocytes. Infection and Immunity. 2003, 71, 4018–4025. [Google Scholar] [CrossRef] [PubMed]
- Castañeda-Ortiz, E.J.; Ueti, M.W.; Camacho-Nuez, M.; Mosqueda, J.J.; Mousel, M.R.; Johnson, W.C.; et al. Association of Anaplasma marginale strain superinfection with infection prevalence within tropical regions. PLoS One. 2015, 10, e0120748. [Google Scholar] [CrossRef] [PubMed]
- Koku, R.; Futse, J.E.; Morrison, J.; Brayton, K.A.; Palmer, G.H.; Noh, S.M. The use of the antigenically variable Major Surface Protein 2 in the establishment of superinfection during natural tick transmission of Anaplasma marginale in Southern Ghana. Infect Immun. 2023, 91, e0050122. [Google Scholar] [CrossRef] [PubMed]
- Singu, V.; Liu, H.; Cheng, C.; Ganta, R.R. Ehrlichia chaffeensis expresses macrophage- and tick cell-specific 28-kilodalton outer membrane proteins. Infect Immun. 2005, 73, 79–87. [Google Scholar] [CrossRef]
- Singu, V.; Peddireddi, L.; Sirigireddy, K.R.; Cheng, C.; Munderloh, U.G.; Ganta, R.R. Unique macrophage and tick cell-specific protein expression from the p28/p30-outer membrane protein multigene locus in Ehrlichia chaffeensis and Ehrlichia canis. Cell Microbiol. 2006, 8, 1475–1487. [Google Scholar] [CrossRef]
- Duan, N.; Ma, X.; Cui, H.; Wang, Z.; Chai, Z.; Yan, J.; et al. Insights into the mechanism regulating the differential expression of the P28-OMP outer membrane proteins in obligatory intracellular pathogen. Emerg Microbes Infect. 2021, 10, 461–471. [Google Scholar] [CrossRef]
- Nyika, A.; Barbet, A.F.; Burridge, M.J.; Mahan, S.M. DNA vaccination with map1 gene followed by protein boost augments protection against challenge with Cowdria ruminantium, the agent of heartwater. Vaccine. 2002, 20, 1215–1225. [Google Scholar] [CrossRef]
- Crocquet-Valdes, P.A.; Thirumalapura, N.R.; Ismail, N.; Yu, X.; Saito, T.B.; Stevenson, H.L.; et al. Immunization with Ehrlichia P28 outer membrane proteins confers protection in a mouse model of ehrlichiosis. Clin Vaccine Immunol. 2011, 18, 2018–2025. [Google Scholar] [CrossRef]
- Budachetri, K.; Lin, M.; Chien, R.C.; Zhang, W.; Brock, G.N.; Rikihisa, Y. Efficacy and immune correlates of OMP-1B and VirB2-4 vaccines for protection of dogs from tick transmission of Ehrlichia chaffeensis. mBio. 2022, 13, e0214022. [Google Scholar] [CrossRef]
- Martin, D.; Rybicki, E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000, 16, 562–563. [Google Scholar] [CrossRef] [PubMed]
- Salminen, M.O.; Carr, J.K.; Burke, D.S.; McCutchan, F.E. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by BOOTSCANning. AIDS Research and Human Retroviruses. 1995, 11, 1423–1425. [Google Scholar] [CrossRef] [PubMed]
- Maynard Smith, J. Analyzing the mosaic structure of genes. Journal of Molecular Evolution. 1992, 34, 126–129. [Google Scholar]
- Posada, D.; Crandall, K.A. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA. 2001, 98, 13757–13762. [Google Scholar] [CrossRef]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-Scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics. 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Weiller, G.F. Phylogenetic profiles: a graphical method for detecting genetic recombinations in homologous sequences. Molecular Biology of Evolution. 1998, 15, 326–335. [Google Scholar] [CrossRef]
- Holmes, E.C.; Worobey, M.; Rambaut, A. Phylogenetic evidence for recombination in Dengue virus. Mol Biol Evol. 1999, 16, 405. [Google Scholar] [CrossRef]
- Lam, H.M.; Ratmann, O.; Boni, M.F. Improved algorithmic complexity for the 3SEQ recombination detection algorithm. Mol Biol Evol. 2018, 35, 247–251. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; et al. UCSF Chimera- a visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004, 25, 1605–1613. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Procter, J.B.; Carstairs, G.M.; Soares, B.; Mourão, K.; Ofoegbu, T.C.; Barton, D.; et al. Alignment of biological sequences with Jalview. Methods Mol Biol. 2021, 2231, 203–224. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




