Submitted:
27 February 2025
Posted:
03 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Retrieval and Analysis of PRRSV Protein Sequence
2.2. Step-by-Step Selection of Epitopes
2.3. Design and Assembly of Candidate Vaccines
2.4. Evaluation of Candidate Vaccines in Physicochemical and Immune Properties
2.5. Prediction and Validation of Molecular Structure for Candidate Vaccines
2.6. Molecular Docking and Dynamic Analysis of Vaccine-TLR Complexes
2.7. Immune Simulation of Candidate Vaccines
2.8. In Silico Cloning of Selected Vaccine Candidate
3. Results
3.1. Extraction of Conserved Fragments from PRRSV Protein Sequences Collected Worldwide
3.2. Prediction and Evaluation of Epitopes for B Cells, CTLs and HTLs
3.3. Construction of Three Candidate Vaccines
3.4. Properties of Candidate Vaccines in Immunology and Physiochemistry
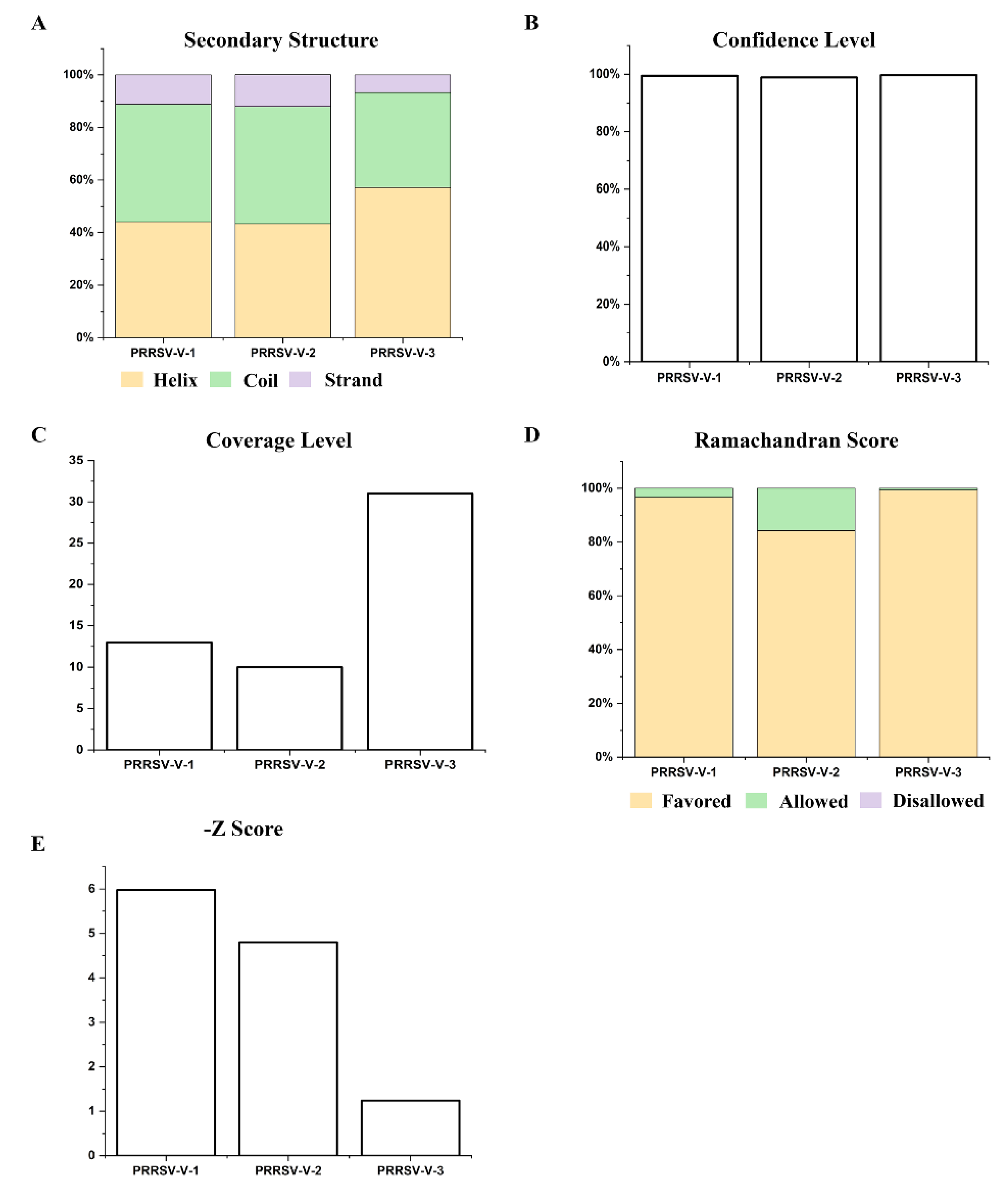
3.5. Prediction and Refinement of Secondary and Tertiary Structure
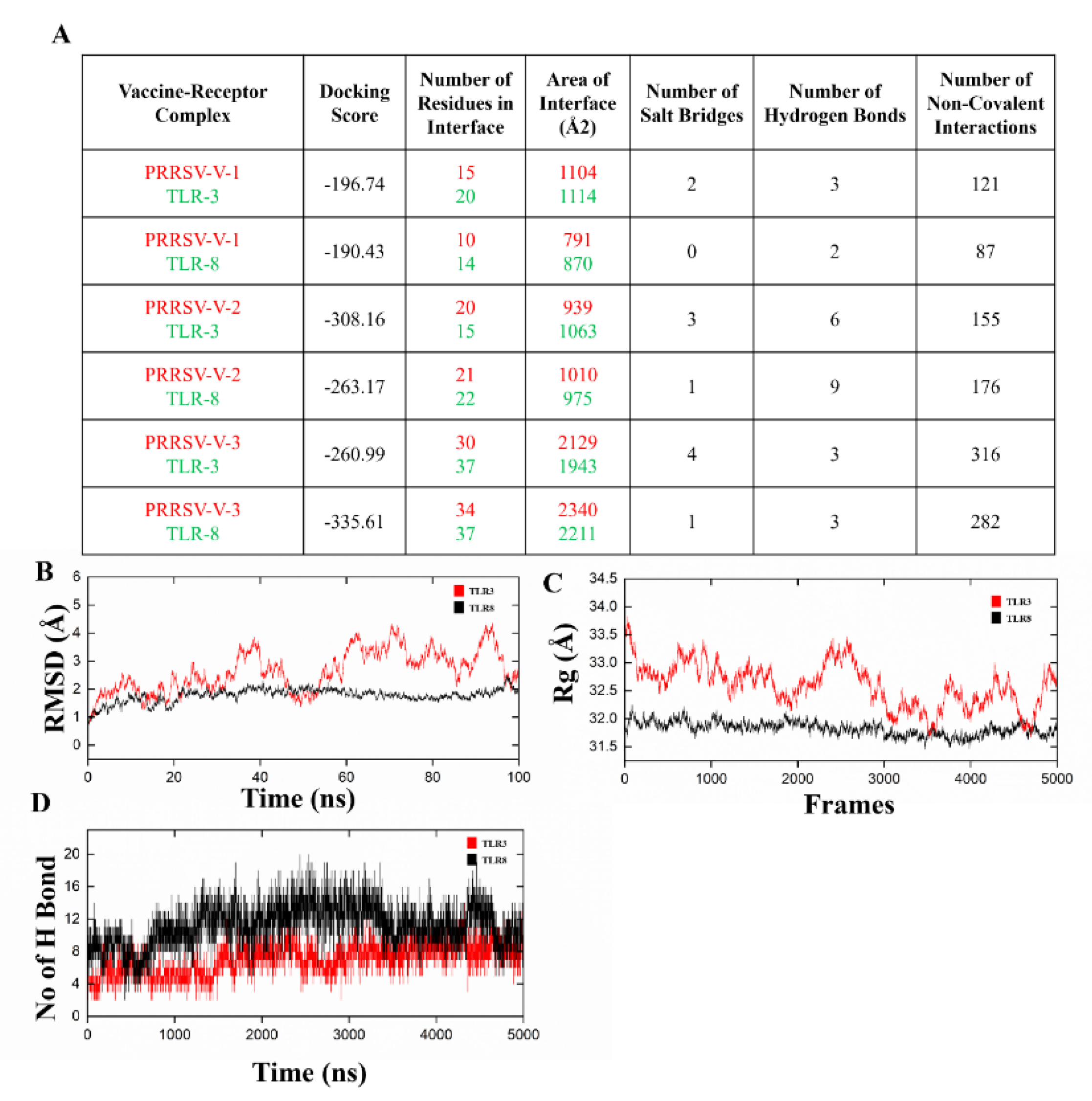
3.6. Molecular Docking and Dynamics Simulation for Complexes of Vaccine-Immune Receptor
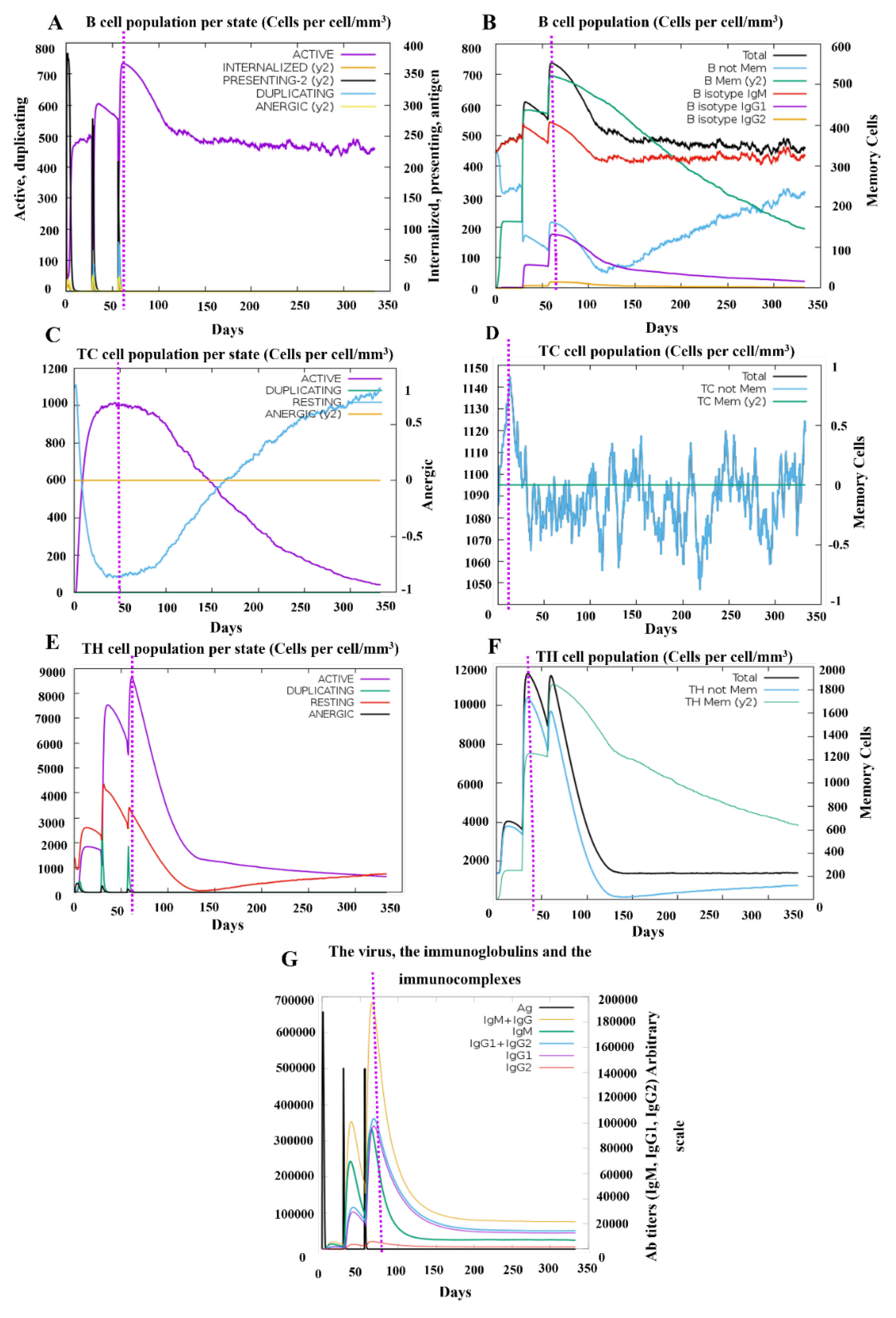
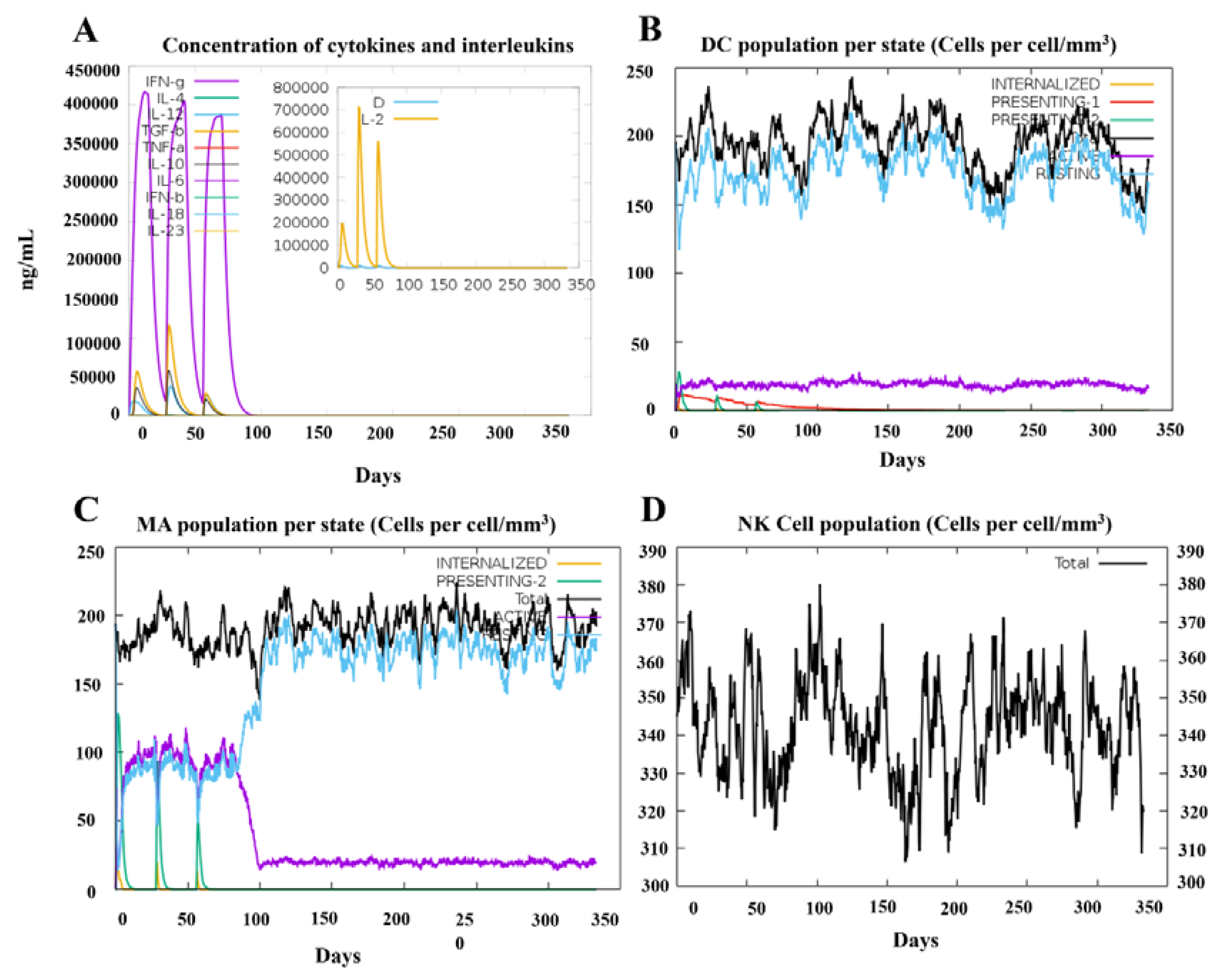
3.9. In Silico Stimulation of Immune Responses
3.10. Codon Optimization of the Final Vaccine Construct
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PRRSV | Porcine Respiratory and Reproductive Syndrome Virus |
| PRRSV-V | Porcine Respiratory and Reproductive Syndrome Virus-vaccine |
| SLA | Swine Leukocyte Antigen |
| HLA | Human Leukocyte Antigen |
References
- Albina, E. Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Veterinary microbiology 1997, 55, 309–316. [Google Scholar] [PubMed]
- Lalonde, C.; Provost, C.; Gagnon, C.A. Whole-genome sequencing of porcine reproductive and respiratory syndrome virus from field clinical samples improves the genomic surveillance of the virus. Journal of clinical microbiology 2020, 58. [Google Scholar] [CrossRef]
- Neumann, E.J.; et al. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. Journal of the American Veterinary Medical Association 2005, 227, 385–392. [Google Scholar]
- Renken, C.; et al. Application of an economic calculator to determine the cost of porcine reproductive and respiratory syndrome at farm-level in 21 pig herds in Germany. Porcine health management 2021, 7, 1–12. [Google Scholar]
- Sun, Q.; et al. Emergence of a novel PRRSV-1 strain in mainland China: A recombinant strain derived from the two commercial modified live viruses Amervac and DV. Frontiers in Veterinary Science 2022, 9, 974743. [Google Scholar]
- Yu, F.; et al. Genomic analysis of porcine reproductive and respiratory syndrome virus 1 revealed extensive recombination and potential introduction events in China. Veterinary Sciences 2022, 9, 450. [Google Scholar]
- Yu, F.; et al. Phylogenetics, genomic recombination, and NSP2 polymorphic patterns of porcine reproductive and respiratory syndrome virus in China and the United States in 2014–2018. Journal of virology 2020, 94. [Google Scholar] [CrossRef]
- Wang, L.-j.; et al. Genomic analysis of a recombinant NADC30-like porcine reproductive and respiratory syndrome virus in China. Virus Genes 2018, 54, 86–97. [Google Scholar]
- Zhao, K.; et al. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. Journal of virology 2015, 89, 10712–10716. [Google Scholar]
- Ruedas-Torres, I.; et al. The jigsaw of PRRSV virulence. Veterinary Microbiology 2021, 260, 109168. [Google Scholar]
- Fang, Y.; Snijder, E.J. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus research 2010, 154, 61–76. [Google Scholar]
- Dea, S.; et al. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Archives of virology 2000, 145, 659–688. [Google Scholar] [PubMed]
- Nan, Y.; et al. Improved vaccine against PRRSV: current progress and future perspective. Frontiers in microbiology 2017, 8, 1635. [Google Scholar]
- Guan, Z.; et al. Secondary Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus (HP-PRRSV2) Infection Augments Inflammatory Responses, Clinical Outcomes, and Pathogen Load in Glaesserella-parasuis-Infected Piglets. Veterinary Sciences 2023, 10, 365. [Google Scholar]
- Zhang, H.; et al. Research Progress on the Development of Porcine Reproductive and Respiratory Syndrome Vaccines. Veterinary Sciences 2023, 10, 491. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Feng, W. Current Status of Porcine Reproductive and Respiratory Syndrome Vaccines. Vaccines 2024, 12, 1387. [Google Scholar] [CrossRef]
- Charerntantanakul, W. Porcine reproductive and respiratory syndrome virus vaccines: Immunogenicity, efficacy and safety aspects. World journal of virology 2012, 1, 23. [Google Scholar]
- Renukaradhya, G.J.; et al. Inactivated and subunit vaccines against porcine reproductive and respiratory syndrome: current status and future direction. Vaccine 2015, 33, 3065–3072. [Google Scholar]
- Chae, C. Commercial PRRS modified-live virus vaccines. Vaccines 2021, 9, 185. [Google Scholar] [CrossRef]
- Lee, M.-A.; et al. Molecular characterization of porcine reproductive and respiratory syndrome virus in Korea from 2018 to 2022. Pathogens 2023, 12, 757. [Google Scholar] [CrossRef]
- Papatsiros, V. Impact of a killed PRRSV vaccine on sow longevity in a PRRSV infected swine herd. Journal of Applied Animal Research 2012, 40, 297–304. [Google Scholar] [CrossRef]
- Kim, H.; et al. The assessment of efficacy of porcine reproductive respiratory syndrome virus inactivated vaccine based on the viral quantity and inactivation methods. Virology journal 2011, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Van Nieuwstadt, A.; et al. Proteins encoded by open reading frames 3 and 4 of the genome of Lelystad virus (Arteriviridae) are structural proteins of the virion. Journal of virology 1996, 70, 4767–4772. [Google Scholar] [CrossRef]
- Cancel-Tirado, S.M.; Evans, R.B.; Yoon, K.-J. Monoclonal antibody analysis of porcine reproductive and respiratory syndrome virus epitopes associated with antibody-dependent enhancement and neutralization of virus infection. Veterinary immunology and immunopathology 2004, 102, 249–262. [Google Scholar] [CrossRef]
- Delputte, P.; et al. Effect of virus-specific antibodies on attachment, internalization and infection of porcine reproductive and respiratory syndrome virus in primary macrophages. Veterinary immunology and immunopathology 2004, 102, 179–188. [Google Scholar] [CrossRef]
- Delputte, P.L.; et al. Porcine arterivirus attachment to the macrophage-specific receptor sialoadhesin is dependent on the sialic acid-binding activity of the N-terminal immunoglobulin domain of sialoadhesin. Journal of virology 2007, 81, 9546–9550. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, M.; et al. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. Journal of virology 2002, 76, 4241–4250. [Google Scholar] [CrossRef]
- Plagemann, P.; Rowland, R.; Faaberg, K. The primary neutralization epitope of porcine respiratory and reproductive syndrome virus strain VR-2332 is located in the middle of the GP5 ectodomain. Archives of virology 2002, 147, 2327–2347. [Google Scholar] [CrossRef]
- Fan, B.; et al. Influence of the amino acid residues at 70 in M protein of porcine reproductive and respiratory syndrome virus on viral neutralization susceptibility to the serum antibody. Virology journal 2016, 13, 1–11. [Google Scholar] [CrossRef]
- Vanhee, M.; et al. A variable region in GP4 of European-type porcine reproductive and respiratory syndrome virus induces neutralizing antibodies against homologous but not heterologous virus strains. Viral immunology 2010, 23, 403–413. [Google Scholar] [CrossRef]
- Costers, S.; et al. GP4 of porcine reproductive and respiratory syndrome virus contains a neutralizing epitope that is susceptible to immunoselection in vitro. Archives of virology 2010, 155, 371–378. [Google Scholar] [CrossRef]
- Costers, S.; et al. GP4-specific neutralizing antibodies might be a driving force in PRRSV evolution. Virus research 2010, 154, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Wissink, E.; et al. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. Journal of virology 2005, 79, 12495–12506. [Google Scholar] [CrossRef] [PubMed]
- Das, P.B.; et al. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. Journal of virology 2010, 84, 1731–1740. [Google Scholar] [CrossRef]
- Meulenberg, J.; et al. Posttranslational processing and identification of a neutralization domain of the GP4 protein encoded by ORF4 of Lelystad virus. Journal of virology 1997, 71, 6061–6067. [Google Scholar] [CrossRef]
- Vashisht, K.; et al. Identification of immunodominant T-cell epitopes present in glycoprotein 5 of the North American genotype of porcine reproductive and respiratory syndrome virus. Vaccine 2008, 26, 4747–4753. [Google Scholar] [CrossRef]
- Díaz, I.; et al. In silico prediction and ex vivo evaluation of potential T-cell epitopes in glycoproteins 4 and 5 and nucleocapsid protein of genotype-I (European) of porcine reproductive and respiratory syndrome virus. Vaccine 2009, 27, 5603–5611. [Google Scholar] [CrossRef]
- Luo, Q.; et al. Research progress on glycoprotein 5 of porcine reproductive and respiratory syndrome virus. Animals 2023, 13, 813. [Google Scholar] [CrossRef]
- Miao, Y.-Q.; et al. RETRACTED ARTICLE: N (6)-adenosine-methyltransferase-14 promotes glioma tumorigenesis by repressing argininosuccinate synthase 1 expression in an m6A-dependent manner. Bioengineered 2022, 13, 1858–1871. [Google Scholar] [CrossRef]
- Gong, H.-R.; et al. Non-neutralizing epitopes shade neutralizing epitopes against omicron in a multiple epitope-based vaccine. ACS Infectious Diseases 2022, 8, 2586–2593. [Google Scholar] [CrossRef] [PubMed]
- Devi, A.; Chaitanya, N.S. In silico designing of multi-epitope vaccine construct against human coronavirus infections. Journal of Biomolecular Structure and Dynamics 2021, 39, 6903–6917. [Google Scholar]
- Ullah, H.; et al. An In Silico Design of a Vaccine against All Serotypes of the Dengue Virus Based on Virtual Screening of B-Cell and T-Cell Epitopes. Biology 2024, 13, 681. [Google Scholar] [CrossRef]
- Yuan, L.; et al. In silico design of a broad-spectrum multiepitope vaccine against influenza virus. International Journal of Biological Macromolecules 2024, 254, 128071. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution 1987, 4, 406–425. [Google Scholar] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar]
- Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research 2004, 32, 1792–1797. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Current protocols in bioinformatics 2003, 1–22. [Google Scholar] [CrossRef]
- Edgar, R.C.; Batzoglou, S. Multiple sequence alignment. Current opinion in structural biology 2006, 16, 368–373. [Google Scholar]
- Oliver, T.; et al. Using reconfigurable hardware to accelerate multiple sequence alignment with ClustalW. Bioinformatics 2005, 21, 3431–3432. [Google Scholar]
- Jespersen, M.C.; et al. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic acids research 2017, 45, W24–W29. [Google Scholar]
- Middleton, D.; et al. New allele frequency database: http://www.allelefrequencies.net. Tissue antigens 2003, 61, 403–407. [Google Scholar] [PubMed]
- Reynisson, B.; et al. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic acids research 2020, 48, W449–W454. [Google Scholar]
- Calis, J.J.; et al. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS computational biology 2013, 9, e1003266. [Google Scholar]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC bioinformatics 2007, 8, 1–7. [Google Scholar]
- Dimitrov, I.; et al. AllerTOP v. 2—a server for in silico prediction of allergens. Journal of molecular modeling 2014, 20, 1–6. [Google Scholar]
- Gupta, S.; et al. In silico approach for predicting toxicity of peptides and proteins. PloS one 2013, 8, e73957. [Google Scholar]
- Dhanda, S.K.; Vir, P.; Raghava, G.P. Designing of interferon-gamma inducing MHC class-II binders. Biology direct 2013, 8, 1–15. [Google Scholar]
- Dhanda, S.K.; et al. Prediction of IL4 Inducing Peptides. Journal of Immunology Research 2013, 2013, 263952. [Google Scholar]
- Fadaka, A.O.; et al. Immunoinformatics design of a novel epitope-based vaccine candidate against dengue virus. Scientific reports 2021, 11, 19707. [Google Scholar]
- Li, W.; et al. Peptide vaccine: progress and challenges. Vaccines 2014, 2, 515–536. [Google Scholar] [CrossRef]
- Jalal, K.; et al. Pan-genome reverse vaccinology approach for the design of multi-epitope vaccine construct against Escherichia albertii. International Journal of Molecular Sciences 2021, 22, 12814. [Google Scholar] [PubMed]
- Dhanushkumar, T.; et al. Structural immunoinformatics approach for rational design of a multi-epitope vaccine against triple negative breast cancer. International Journal of Biological Macromolecules 2023, 243, 125209. [Google Scholar]
- Ghaffari-Nazari, H.; et al. Improving multi-epitope long peptide vaccine potency by using a strategy that enhances CD4+ T help in BALB/c mice. PloS one 2015, 10, e0142563. [Google Scholar]
- Gasteiger, E.; et al. Protein Analysis Tools on the ExPASy Server 571 571 From: The Proteomics Protocols Handbook Protein Identification and Analysis Tools on the ExPASy Server. The Proteomics Protocols Handbook 2019, 571–607. [Google Scholar]
- Magnan, C.N.; Randall, A.; Baldi, P. SOLpro: accurate sequence-based prediction of protein solubility. Bioinformatics 2009, 25, 2200–2207. [Google Scholar]
- McGuffin, L.J.; Bryson, K.; Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [PubMed]
- Kelley, L.A.; et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nature protocols 2015, 10, 845–858. [Google Scholar] [PubMed]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic acids research 2013, 41, W384–W388. [Google Scholar]
- Wiederstein, M.; Sippl, M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic acids research 2007, 35 (Suppl. 2), W407–W410. [Google Scholar] [CrossRef]
- Laskowski, R.A.; et al. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of applied crystallography 1993, 26, 283–291. [Google Scholar]
- Liu, D.; Chen, Y. Epitope screening and vaccine molecule design of PRRSV GP3 and GP5 protein based on immunoinformatics. Journal of Cellular and Molecular Medicine 2024, 28, e18103. [Google Scholar] [CrossRef] [PubMed]
- Ullah, M.A.; Sarkar, B.; Islam, S.S. Exploiting the reverse vaccinology approach to design novel subunit vaccines against Ebola virus. Immunobiology 2020, 225, 151949. [Google Scholar] [CrossRef]
- Yan, Y.; et al. The HDOCK server for integrated protein–protein docking. Nature protocols 2020, 15, 1829–1852. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; et al. PDBsum: Structural summaries of PDB entries. Protein science 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Lovell, S.C.; et al. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins Structure Function and Bioinformatics 2003, 50, 437–450. [Google Scholar] [CrossRef]
- Case, D.; et al. The FF14SB force field. Amber 2014, 14, 29–31. [Google Scholar]
- Maier, J.A.; et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. Journal of chemical theory and computation 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Wang, J.; et al. Development and testing of a general amber force field. Journal of computational chemistry 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Kräutler, V.; Van Gunsteren, W.F.; Hünenberger, P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. Journal of computational chemistry 2001, 22, 501–508. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. Journal of chemical theory and computation 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr 2002, 40, 82–92. [Google Scholar]
- Rapin, N.; et al. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PloS one 2010, 5, e9862. [Google Scholar] [CrossRef] [PubMed]
- Grote, A.; et al. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic acids research 2005, 33 (Suppl. 2), W526–W531. [Google Scholar] [CrossRef]
- Ullah, H.; et al. An In Silico Design of a Vaccine against All Serotypes of the Dengue Virus Based on Virtual Screening of B-Cell and T-Cell Epitopes. Biology 2024, 13. [Google Scholar] [CrossRef] [PubMed]
| Genotype | protein | Start -end |
epitopes | length | Antigenicity Score |
Allergenicity /Toxicity |
Intra/Inter Conservancy |
|---|---|---|---|---|---|---|---|
| PRRSV1 | ORF2A | 57-66 | APRYSVRALP | 10 | 0.8784 | Non/Non | 75.00% /20.00% |
| PRRSV2 | GP3 | 38-45 | LVRGNFSF | 8 | 1.7037 | Non/Non | 100.00% /33.33% |
| PRRSV2 | GP4 | 44-57 | QHTQQHHLVIDHIR | 14 | 0.5457 | Non/Non | 100.00% /33.33% |
| PRRSV1 | GP5 | 49-61 | LNGTDWLDKRFDW | 13 | 1.7333 | Non/Non | 75.00% /20.00% |
| PRRSV2 | M | 131-144 | RKPGLTSVNGTLV | 13 | 0.8659 | Non/Non | 100.00% /33.33% |
| PRRSV1 | N | 6-13 | HFPLAAED | 8 | 0.9417 | Non/Non | 66.00% /22.22% |
| Genotype | protein | SLA Allele | Epitope | Antigenicity /Immunogenicity |
Allergenicity /Toxicity |
Intra/Inter Conservancy |
|---|---|---|---|---|---|---|
| PRRSV1 | ORF1a | 1*1101 | AALTGRTL | 1.2071 /0.14994 |
Non/Non | 100.00% /25.00% |
| PRRSV1 | ORF1a | 3*0501 | HQKPIAYANL | 1.1782 /0.1123 |
Non/Non | 100.00% /20.00% |
| PRRSV2 | ORF2b | 3*0701 | TRARHAIF | 1.0977 /0.31383 |
Non/Non | 95.24% /25.00% |
| PRRSV2 | GP2a | 3*0401 | SVRALPFTL | 1.5046 /0.15641 |
Non/Non | 75.00% /22.22% |
| PRRSV1 | GP2b | 2*0101 | IFLAILFGF | 0.7123 /0.27353 |
Non/Non | 75.00% /22.22% |
| PRRSV2 | GP3 | 2*0101 | YAWLAFLSF | 1.1215 /0.10302 |
Non/Non | 100.00% /22.22% |
| PRRSV2 | GP4 | 1*0702 | SACVNFTDY | 1.6035 /0.17366 |
Non/Non | 66.67% /22.22% |
| PRRSV2 | GP5 | 3*0101 | TRYTNFLL | 1.2753 /0.1324 |
Non/Non | 66.67% /25.00% |
| PRRSV1 | M | 2*1201 | FSITYTPII | 1.4228 /0.1836 |
Non/Non | 66.67% /22.22% |
| PRRSV1 | N | 2*1101 | FPLATEDDVRHHF | 0.4645 /0.34939 |
Non/Non | 50.00% /38.46% |
| Genotype | Protein | HLA- Allele | Epitope | Antigenicity | Allergenicity/toxicity | IL-4/IFN- γ induction | Intra/Inter Conservancy |
|---|---|---|---|---|---|---|---|
| PRRSV1 | ORF1 | DPA1*02:01 DPB1*05:01 |
GVAPAVRIAERYRGR | 0.9596 | Non/Non | Inducer /Inducer |
100.00% /13.33% |
| PRRSV1 | ORF | DRB4*01:04 | KPIAYANLDEKKISA | 0.9123 | Non/Non | Inducer /Inducer |
100.00% /13.33% |
| PRRSV2 | ORF1b | DRB1*12:04 | CLGDFKQLHPVGFDS | 1.2326 | Non/Non | Inducer /Inducer |
95.24% /20.00% |
| PRRSV2 | GP2a | DPA1*02:01 DPB1*04:01 |
LSFASDWFAPRYSVR | 0.5461 | Non/Non | Inducer /Inducer |
75.00% /20.00% |
| PRRSV2 | GP2b | DRB1*04:04 | VFCIRLVCSAIHRSR | 1.3347 | Non/Non | Inducer /Inducer |
100.00% /20.00% |
| PRRSV2 | GP3 | DPA1*02:01 DPB1*04:01 |
NWFHLEWLRPFFSSW | 0.4583 | Non/Non | Inducer /Inducer |
75.00% /20.00% |
| PRRSV1 | GP4 | DRB5*01:03 | ACVNFTDYVAHVTQH | 1.0257 | Non/Non | Inducer /Inducer |
75.00% /13.33% |
| PRRSV2 | GP5 | DPA1*01:03 DPB1*01:01 |
WRYSCTRYTNFLLDT | 0.5114 | Non/Non | Inducer /Inducer |
100.00% /13.33% |
| PRRSV1 | M | DPA1*02:01 DPB1*02:01 |
LAFSITYTPIIYALK | 1.4017 | Non/Non | Inducer /Inducer |
100.00% /13.33% |
| PRRSV2 | N | DQA1*01:01 DQB1*02:01 |
PHFPLATEDDVRHHF | 0.4807 | Non/Non | Inducer /Inducer |
66.67% /26.67% |
| Parameters | PRRSV-V-1 vaccine | PRRSV-V-2 vaccine | PRRSV-V-3 vaccine |
|---|---|---|---|
| No. of amino acids | 531 | 448 | 551 |
| Molecular weight | 57526.93 | 49446.89 | 60733.93 |
| Instability Index | 22.04 | 25.72 | 28.66 |
| Aliphatic index | 81.02 | 80.91 | 77.04 |
| Half-life | 30 h (mammalian reticulocytes, in vitro). >20 h (yeast, in vivo). >10 h (Escherichia coli, in vivo) |
30 h (mammalian reticulocytes, in vitro). >20 h (yeast, in vivo). >10 h (Escherichia coli, in vivo) |
30 h (mammalian reticulocytes, in vitro). >20 h (yeast, in vivo). >10 h (Escherichia coli, in vivo) |
| Solubility | 0.979905 | 0.975094 | 0.986037 |
| GRAVY | -0.054 | -0.191 | -0.249 |
| Theoretical pI | 9.12 | 9.82 | 9.17 |
| Antigenicity | 0.61 | 0.68 | 0.63 |
| Allergenicity | Non-Allergenic | Non-Allergenic | Non-Allergenic |
| Toxicity | Non-Toxic | Non-Toxic | Non-Toxic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





