Submitted:
01 March 2025
Posted:
03 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. RNA-DNA Differences: A Nexus Linking Oxidative Stress and Genomic Instability
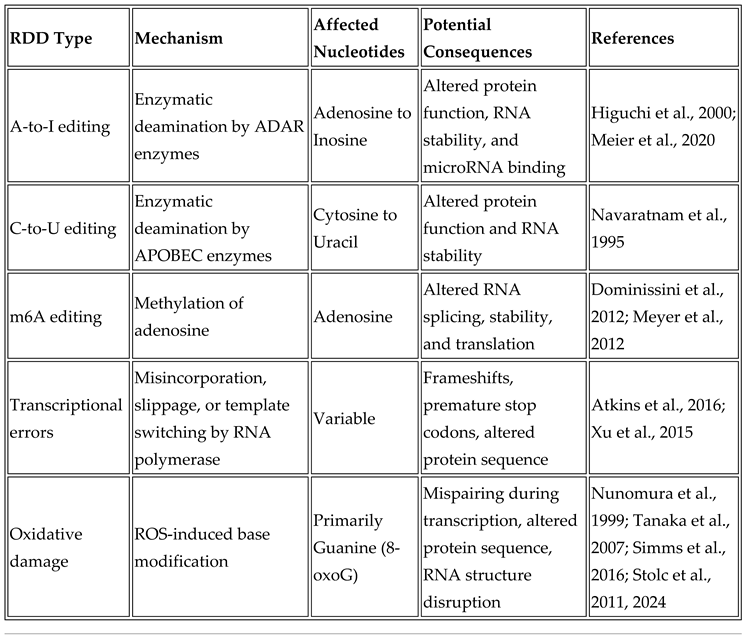
2.2. Mechanisms of RNA-DNA Differences
2.3. Oxidative Stress and the Formation of RDDs
3. Functional Consequences of RDDs
- Protein dysfunction: By altering codons, RDDs can lead to incorrect amino acid incorporation during translation, producing misfolded or non-functional proteins. Confirmed by mass spectrometry sequencing, these changes represent bona fide mutations due to permanent amino acid sequence alterations. For instance, oxidation-induced mutation of tryptophan codons (UGG) to stop codons (UAG) results in truncated proteins, often unstable and prone to aggregation (Sorrentino et al., 2018). These effects are particularly detrimental in neurodegenerative diseases, where protein aggregation contributes to synaptic dysfunction and cell death (Wheeler et al., 2024).
- Genomic instability: Beyond protein dysfunction, RDDs can impact DNA repair pathways, telomere maintenance, and epigenetic modifications. RDDs could affect the expression or function of DNA repair enzymes, compromising DNA damage repair. Similarly, RDDs could influence telomere length or the expression of telomere-associated proteins, potentially contributing to cellular senescence. Additionally, RDDs may alter the expression or activity of epigenetic modifiers, leading to changes in gene expression patterns. Furthermore, large-scale genomic instability can stem from oxidative damage, including chromosomal rearrangements arising from defects in repairing oxidative DNA damage (particularly abasic sites) or from nucleotide pool imbalances (Kumar et al., 2011). These imbalances can lead to errors in DNA replication and repair, further contributing to genomic instability. Therefore, large-scale genomic instability should be viewed as a potential consequence of oxidative stress and impaired DNA repair (Ragu et al., 2007; Iraqui et al., 2009; Degtyareva et al., 2008; Evert et al., 2004; Kumar et al., 2010).
- Immune responses: Disruption of G4 structures by RDDs can further exacerbate these consequences. G4s are crucial for telomere integrity, and damage to telomeric G4s can lead to telomere shortening and genomic instability. Oxidative damage at telomeres, particularly the conversion of guanine to 8-oxoG, can disrupt the protective G-quadruplex structures, contributing to telomere shortening and genomic instability. This process is further exacerbated by the impaired excision of 8-oxoG by OGG1 due to the unique secondary structures at telomeres (Poetsch A. R. (2020). Additionally, G4s in gene promoters can regulate gene expression, and their disruption can lead to altered transcription and translation.
-
Beyond protein synthesis, RDDs can:
- Inhibit RNase P activity: RDDs in tRNA can alter their structure and function, potentially inhibiting RNase P activity, a ribozyme essential for tRNA maturation. This inhibition can disrupt tRNA processing and protein synthesis, leading to cellular dysfunction (Altman & Stolc, 1997; Samanta et al., 2006). Additionally, the catalytic RNA component of RNase P (Guerrier-Takada et al., 1983; Jarrous & Liu, 2023) can be oxidized, and RDDs in this RNA can further inhibit its catalytic activity on pre-tRNAs.
- Disrupt other RNP complexes: RDDs in RNA components of various ribonucleoprotein (RNP) complexes can potently inhibit essential cellular processes. For example, RDDs in the signal recognition particle (SRP) RNA can impair its ability to target proteins to the endoplasmic reticulum (ER) for secretion, and ER stress is closely linked to ROS production as part of the unfolded protein response (Jiang et al., 2020). Similarly, RDDs in spliceosome RNA components can disrupt mRNA splicing, leading to aberrant protein production (Cech, 2018).
- Alter microRNA binding: RDDs in mRNA can affect microRNA binding sites, leading to gene expression dysregulation and contributing to disease (Vaghf et al., 2022).
- Change RNA localization: RDDs might influence RNA structure-based interactions required for trafficking and localization, affecting its functions (Cui et al., 2022).
- Trigger immune response: RDDs can be recognized as "non-self" by cellular sensors, triggering innate immune responses and contributing to inflammation (Yuan et al., 2023). For example, RDDs can affect Y-RNAs-based stress response, immune activation, and genomic stability (Boccitto & Wolin, 2019).
- Defective chromosome replication: Telomerase, a ribonucleoprotein enzyme complex, is crucial for maintaining telomere integrity. Telomeres are protective caps at chromosome ends that safeguard genomic stability. The RNA subunit of telomerase, TERC, provides the template for telomere extension. However, TERC is highly susceptible to oxidative damage caused by reactive oxygen species (ROS), which impairs telomerase activity and accelerates telomere shortening. This, coupled with the inherent vulnerability of telomeric DNA to oxidative stress, promotes genomic instability and cellular senescence, a state of irreversible cell cycle arrest.
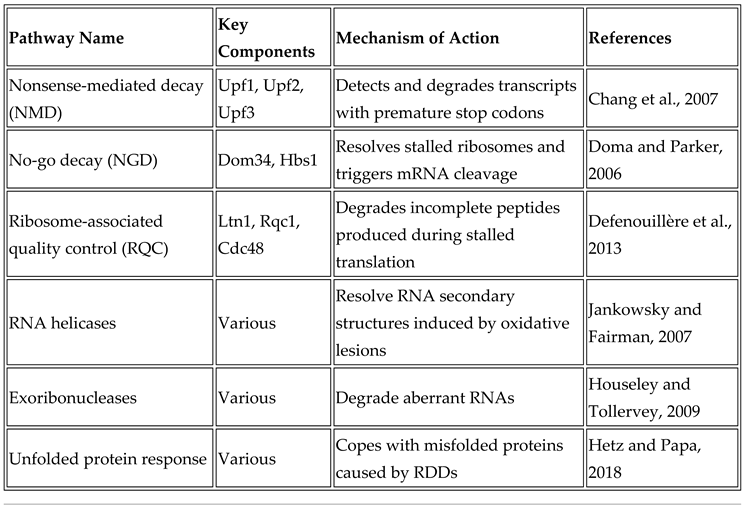
4. Cellular Mechanisms to Counteract RDDs
- Nonsense-mediated decay (NMD): NMD is a critical mRNA surveillance mechanism that identifies and degrades transcripts containing premature termination codons (PTCs), which can arise from mutations or RDDs. By eliminating these faulty mRNAs, NMD prevents truncated, potentially harmful protein synthesis. The process involves PTC recognition, SURF complex (SMG-1, UPF1, eRF1, and eRF3) assembly, and recruitment of RNA degradation machinery (Behera et al., 2024).
- No-go decay (NGD): NGD addresses ribosomal stalling during translation, which can result from obstacles like strong RNA secondary structures or oxidative lesions. When a ribosome stalls, NGD detects it and initiates endonucleolytic cleavage near the stall site, followed by mRNA fragment degradation by exonucleases (Yan et al., 2019).
- Ribosome-associated quality control (RQC): RQC manages incomplete nascent peptide degradation resulting from stalled translation. Upon ribosomal stalling, RQC facilitates ribosomal subunit dissociation and targets the incomplete polypeptide for ubiquitination and proteasomal degradation, preventing defective protein accumulation (Yan et al., 2019).
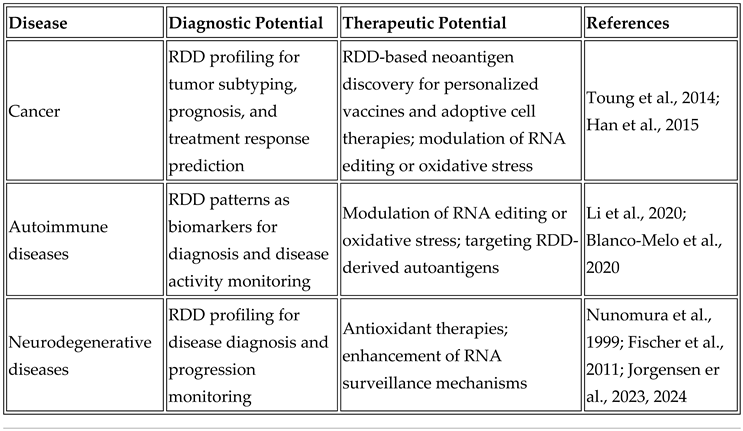
5. The Clinical Potential of RNA-DNA Differences: Neoantigens in Cancer Immunotherapy and Autoantigens in Autoimmune Diseases
6. RDDs as Neoantigens in Cancer Immunotherapy
7. RDDs as Autoantigens in Autoimmune Diseases
8. Balancing Therapeutic Potential and Pathogenic Risks
9. Adaptive and Clinical Implications
10. Biochemical Explanation for GlyNAC's Effectiveness in Reducing ROS
11. Therapeutic Insights
12. Hypometabolism as a Therapeutic Intervention
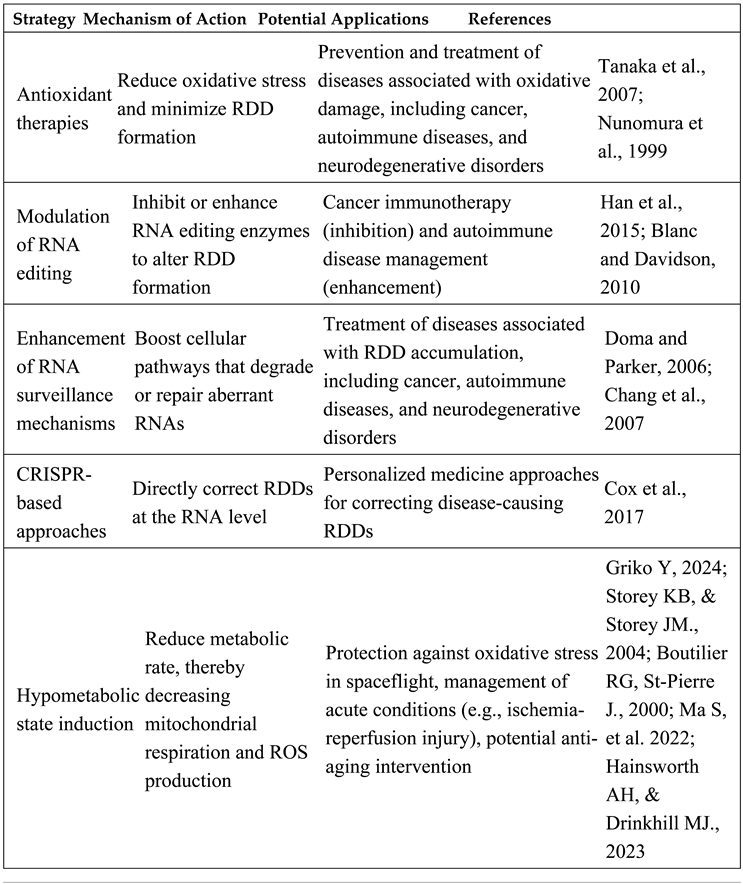
13. Mitigation Strategies for RNA-DNA Differences (RDDs)
- Oxidative Stress Reduction: Antioxidant therapies such as N-acetylcysteine (NAC), GlyNAC, quercetin, and vitamin E can enhance cellular defenses against ROS. Modulating gasotransmitter levels (CO and H₂S) offers additional control by preventing excess ROS production linked to mitochondrial dysfunction.
- Enhanced RNA and DNA Repair: Strengthening cellular pathways like nonsense-mediated decay (NMD), no-go decay (NGD), and ribosome-associated quality control (RQC) ensures more efficient degradation of aberrant RNA molecules. Emerging RNA repair technologies modeled on DNA repair pathways, such as CRISPR-Cas13-based editing, present promising tools for directly correcting RDDs.
- Advanced Detection and Quantification: High-throughput sequencing and mass spectrometry, coupled with bioinformatics tools like PUFFIN, allow for precise mapping and monitoring of RDDs, providing insights into their formation and functional consequences.
- Therapeutic Modulation of RNA Editing: Targeted modulation of ADAR and APOBEC enzymes can either inhibit harmful RNA editing in cancer or enhance beneficial editing in other contexts, offering potential therapeutic leverage.
- Disease-Specific Approaches: In cancer, RDD-derived neoantigens present opportunities for personalized immunotherapies, including mRNA vaccines and CAR T-cell therapies. For autoimmune diseases, reducing oxidative stress and modulating RNA editing pathways could mitigate autoantigenic RDDs.
- Personalized Medicine and Spaceflight Applications: Personalized RDD profiles can inform tailored therapies, while spaceflight-specific interventions, such as environmental controls and radiation shielding, address unique oxidative stress challenges.
14. Conclusions
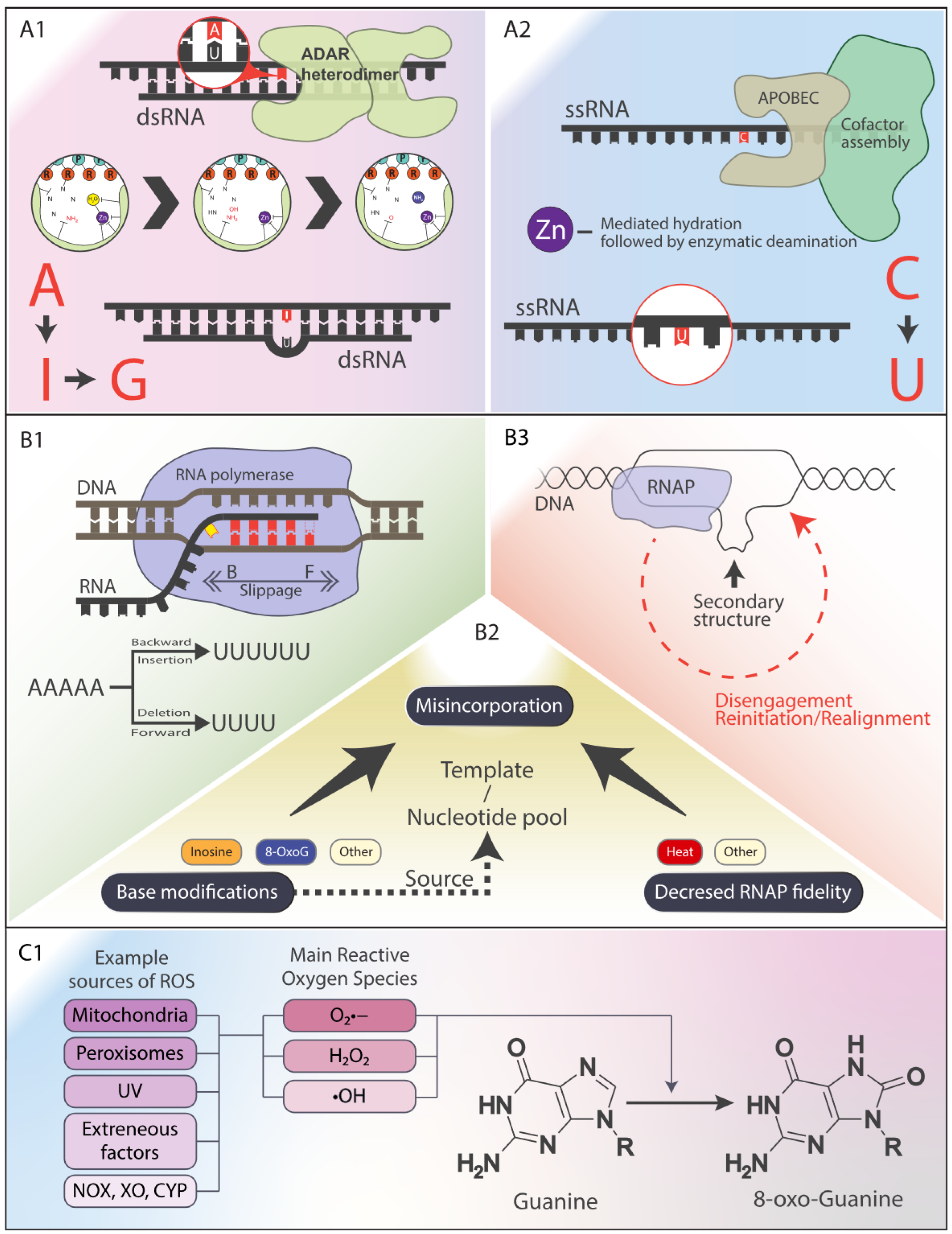
- RNA-DNA Differences (RDDs) arise from diverse mechanisms that introduce sequence discrepancies between genomic DNA and transcribed RNA. This figure highlights key contributors to RDD formation:
-
(A) Enzymatic RNA Editing: Post-transcriptional modifications that alter RNA bases.
- (A1) ADAR Enzymes: Adenosine Deaminases Acting on RNA (ADARs) catalyze the deamination of adenosine (A) to inosine (I), which is interpreted as guanine (G) during translation, leading to A-to-G transitions. ADAR editing is critical for transcriptome diversity, particularly in repetitive elements like Alu sequences, and influences immune function and neural processes. Dysregulation is linked to diseases such as cancer and neurological disorders.
- (A2) APOBEC Enzymes: While primarily involved in DNA editing and antiviral defense, certain Apolipoprotein B mRNA Editing Catalytic Polypeptide-like (APOBEC) enzymes can catalyze cytosine (C) to uracil (U) deamination in RNA. The specific functions and targets of APOBEC-mediated RNA editing remain under investigation.
-
(B) Transcriptional Errors: Mistakes occurring during RNA synthesis.
- (B1) Polymerase Slippage: RNA polymerase can slip at homopolymeric runs (e.g., AAAAA), leading to insertions or deletions (indels) in the transcript. This is more frequent in repetitive genomic regions.
-
(B2) Misincorporation: Incorrect nucleotide incorporation can arise due to:
- ○
- Modified bases in DNA or the nucleotide pool (e.g., 8-oxoG mispairing).
- ○
- Reduced polymerase fidelity caused by environmental stressors (e.g., high temperature) or mutations.
- ○
- Although proofreading and RNA surveillance mechanisms correct most errors, uncorrected misincorporation can contribute to RDD formation.
- (B3) Template Switching: RNA polymerase may switch templates when encountering DNA secondary structures (e.g., hairpins, G-quadruplexes) or DNA lesions, producing chimeric RNA molecules. This mechanism is also exploited by some viruses (e.g., retroviruses) to enhance genetic diversity.
-
(C) Oxidative Damage: ROS-induced RNA modifications.
- (C1) 8-oxoG and ROS: 8-oxoguanine (8-oxoG) is a major oxidative lesion generated by reactive oxygen species (ROS) from mitochondrial respiration, inflammation, and environmental stress (e.g., radiation, pollutants). During transcription, 8-oxoG mispairs with adenine (A), leading to G-to-T transversions in RNA. These oxidative modifications contribute to genomic and transcriptomic instability, potentially driving mutagenesis and disease.
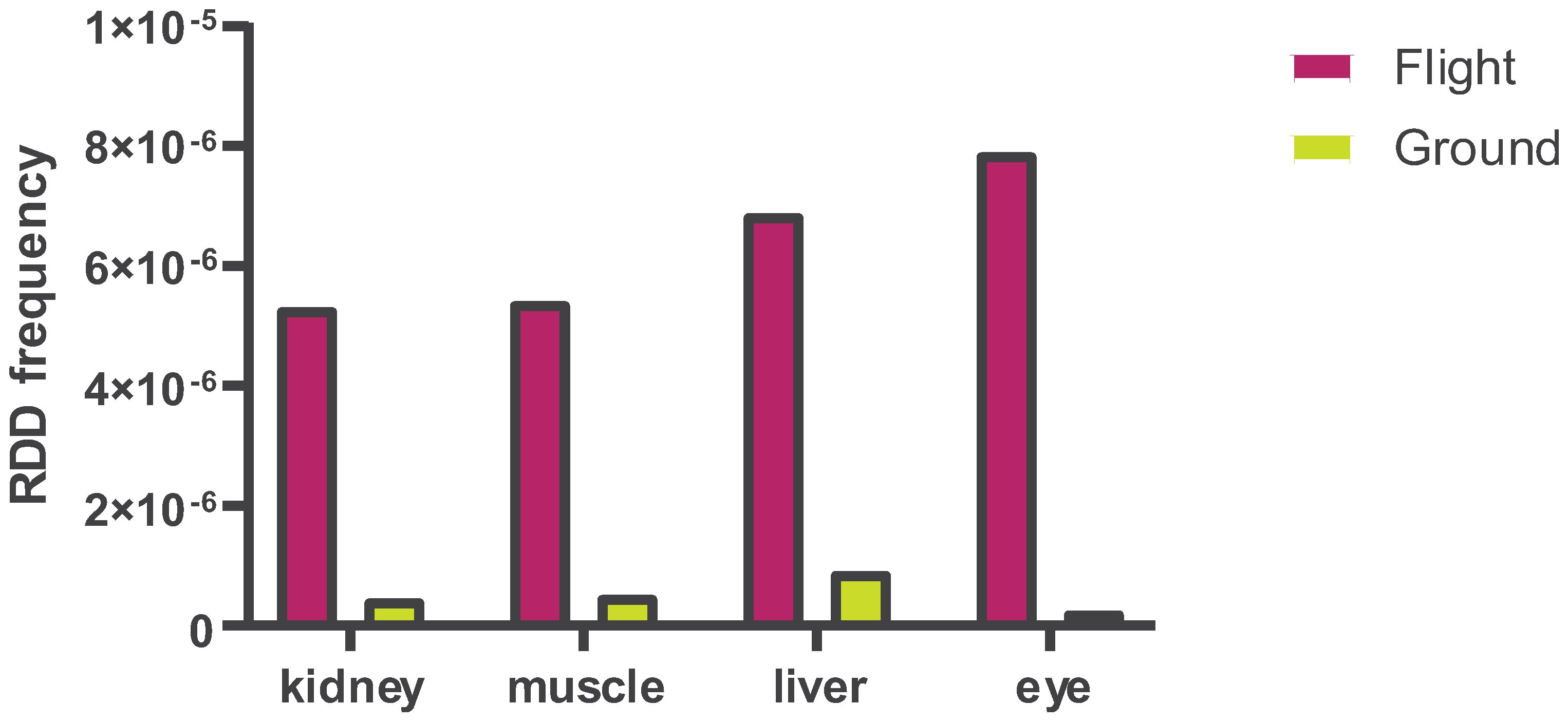
- (A) RDD Frequency Across Tissues: This panel compares RNA-DNA difference (RDD) frequencies in four tissues – kidney, muscle, liver, and eye – from mice exposed to spaceflight (ISS) for 37 days versus ground controls (Earth). Paired bar graphs represent each tissue type. The right bar in each pair shows the mean RDD frequency in terrestrial control samples, while the left bar shows the mean RDD frequency in matched samples from ISS-flown mice.
- This figure depicts the self-perpetuating cycle linking oxidative damage to RNase MRP and RNase P RNA with mitochondrial dysfunction and increased reactive oxygen species (ROS) production, ultimately driving further RNA oxidation and genomic instability.
-
(1) Impaired RNA Function:
- RNase MRP: Oxidative modifications to RNase MRP RNA compromise its crucial roles in mitochondrial DNA replication and pre-rRNA processing. This leads to impaired mitochondrial genome maintenance and ribosome biogenesis, reducing mitochondrial protein synthesis and ultimately causing defects in oxidative phosphorylation and diminished ATP production.
- RNase P: Oxidative damage to RNase P RNA disrupts tRNA processing, resulting in an accumulation of improperly matured tRNAs and subsequent translation defects. This further exacerbates cellular stress and compromises protein synthesis efficiency.
-
(2) Mitochondrial Dysfunction and ROS Amplification:
- The combined effects of impaired RNase MRP and RNase P function lead to mitochondrial dysfunction, characterized by excessive ROS production. This heightened ROS environment further targets RNase MRP and RNase P RNA, amplifying oxidative RNA damage and establishing a vicious cycle.
- The increased ROS levels drive the formation of additional RNA-DNA differences (RDDs), reinforcing mitochondrial instability and perpetuating a state of genomic and transcriptomic instability.
-
(3) Consequences and Broader Implications:
- This feedback loop extends beyond mitochondrial dysfunction, as the continual accumulation of RDDs contributes to a decline in overall genomic stability, impaired translation fidelity, and increased cellular vulnerability to oxidative stress.
- Over time, this cycle accelerates genomic entropy, underscoring the fundamental biochemical constraint that efficient energy production in the mitochondria is inherently linked to an inevitable increase in ROS-induced damage.
- This interplay between oxidative stress and RNA modification highlights the fragility of cellular systems and the persistent challenge of maintaining genomic and mitochondrial integrity in metabolically active environments. It also emphasizes the potential for therapeutic interventions targeting mitochondrial health and RNA quality control mechanisms to mitigate the detrimental effects of this vicious cycle.
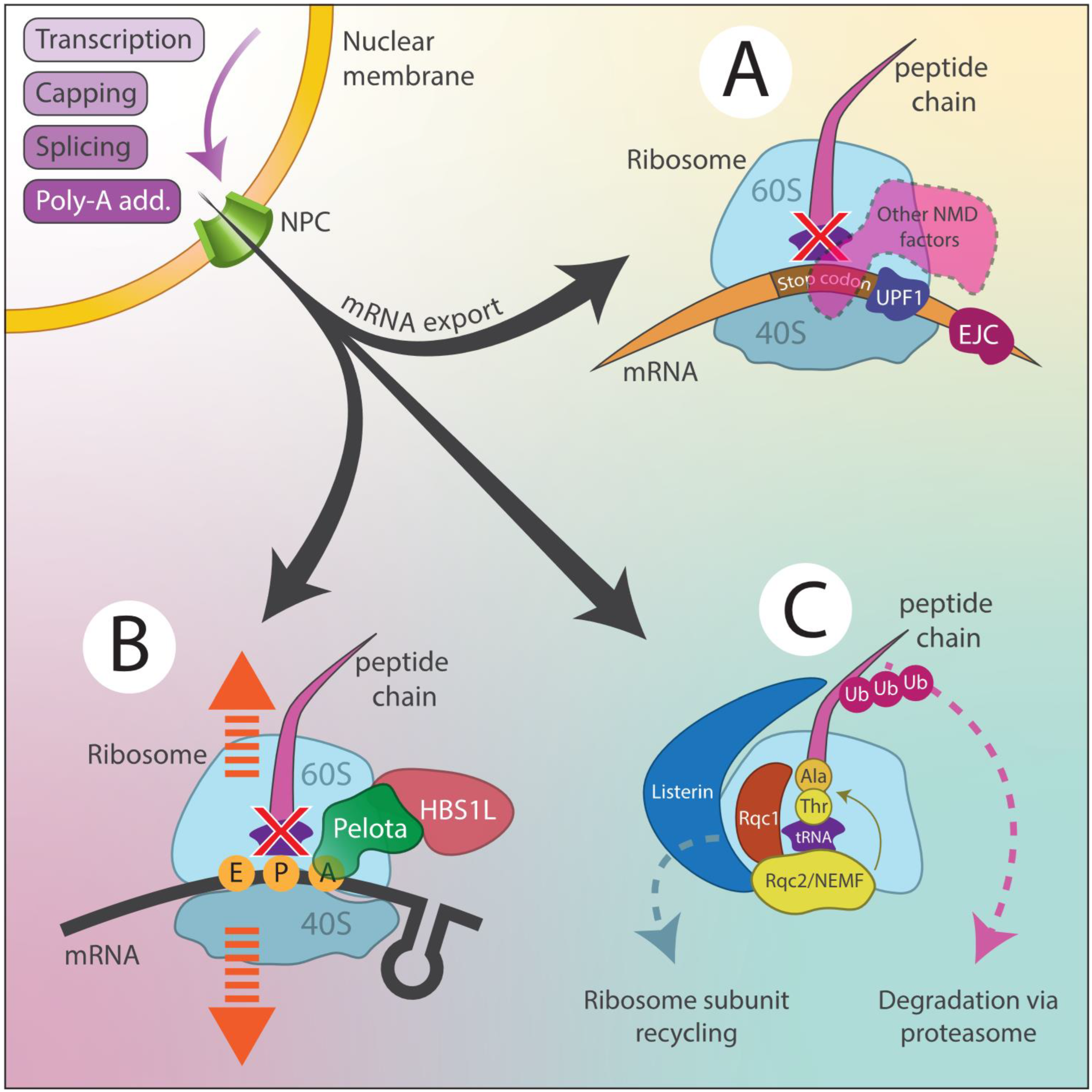
- This figure illustrates the key quality control pathways that recognize and degrade faulty mRNAs or proteins arising from RNA-DNA differences (RDDs), ensuring proper cellular function.
-
(A) Nonsense-Mediated Decay (NMD)
- A mRNA surveillance pathway that prevents the accumulation of truncated, potentially toxic proteins.
-
UPF1, a central RNA helicase, recognizes premature stop codons, triggering a cascade of events:
- ○
- Decapping (removal of the 5′ cap).
- ○
- Deadenylation (removal of the 3′ poly(A) tail).
- ○
- Exonucleolytic degradation by the exosome and XRN1, ensuring faulty transcripts are eliminated.
-
(B) No-Go Dcay (NGD)
- A response to ribosome stalling caused by structural obstacles, damaged codons, or defective RNA.
- Dom34 (Pelota) and Hbs1 recognize the stalled ribosome and cleave the problematic mRNA near the stall site.
- The truncated mRNA fragments are degraded by exonucleases such as XRN1 and the exosome complex.
- The stalled ribosome is recycled, preventing translation bottlenecks.
-
(C) Ribosome-Associated Quality Control (RQC)
- Ensures degradation of incomplete polypeptides that stall on ribosomes.
- Listerin (LTN1), an E3 ubiquitin ligase, ubiquitinates the stalled polypeptide, marking it for proteasomal degradation.
- NEMF (Rqc2 homolog) facilitates CAT tail (C-terminal alanine-threonine extension) addition, signaling faulty peptides for disposal.
- ANKZF1 (Vms1 homolog, not displayed) cleaves stalled peptides and aids in ribosome recycling.
References
- Ahmed, S. , Passos, J. F., & Birket, M. J. (2008). Telomere dysfunction and oxidative stress: Mechanisms of accelerated aging? Antioxidants & Redox Signaling, 10(2), 363–372. [CrossRef]
- Amariei C, Machné R, Stolc V, Soga T, Tomita M, Murray DB. Time resolved DNA occupancy dynamics during the respiratory oscillation uncover a global reset point in the yeast growth program. Microb Cell 2014;1(9):279-288. [CrossRef]
- Ames, B. N. , Cathcart, R., Schwiers, E., & Hochstein, P. (1981). Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: a hypothesis. Proceedings of the National Academy of Sciences of the United States of America, 78(11), 6858–6862. [CrossRef]
- Amin M, Brooks BR. The oxidation of the [4Fe-4S] cluster of DNA primase alters the binding energies with DNA and RNA primers. Biophys J. 2024;123(12):1648-1653. [CrossRef]
- Amrein K, Oudemans-van Straaten HM, Berger MM. Vitamin therapy in critically ill patients: focus on thiamine, vitamin C, and vitamin D. Intensive Care Med. 2018;44(11):1940-1944. [CrossRef]
- Andersen et al., 2003: Andersen, P. M., & Al-Chalabi, A. (2011). Clinical genetics of amyotrophic lateral sclerosis: What do we really know? Nature Reviews Neurology, 7(11), 603-615. [CrossRef]
- Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016;44(15):7007-7078. [CrossRef]
- Back P, Braeckman BP, Matthijssens F. ROS in aging Caenorhabditis elegans: damage or signaling? Oxid Med Cell Longev. 2012;2012:608478. [CrossRef]
- Bass, B. L. (2002). RNA editing by adenosine deaminases that act on RNA. Annual Review of Biochemistry, 71(1), 817-846. [CrossRef]
- Behera A, Panigrahi GK, Sahoo A. Nonsense-Mediated mRNA Decay in Human Health and Diseases: Current Understanding, Regulatory Mechanisms and Future Perspectives. Mol Biotechnol. Published online September 12, 2024. [CrossRef]
- Barger JL, Brand MD, Barnes BM, Boyer BB. Tissue-specific depression of mitochondrial proton leak and substrate oxidation in hibernating arctic ground squirrels. Am J Physiol Regul Integr Comp Physiol. 2003;284(5):R1306-R1313. [CrossRef]
- Bergendi, L. , Benes, L., Durackova, Z., & Ferencik, M. (1999). Chemistry, physiology and pathology of free radicals. Life Sciences, 65(18-19), 1865-1874. [CrossRef]
- Barnes, PJ. Oxidative Stress in Chronic Obstructive Pulmonary Disease. Antioxidants (Basel). 2022;11(5):965. [CrossRef]
- Baird BJ, Dickey JS, Nakamura AJ, et al. Hypothermia postpones DNA damage repair in irradiated cells and protects against cell killing. Mutat Res. 2011;711(1-2):142-149. [CrossRef]
- Blanc, V. , & Davidson, N. O. (2003). APOBEC-1-mediated RNA editing. Wiley Online Library. [CrossRef]
- Blanco-Melo, D. , et al. (2020). RNA modifications in immune regulation and autoimmunity. Nature Immunology. 21(4), 393–402. [CrossRef]
- Blasco, M. A. (2005). Telomeres and human disease: Ageing, cancer and beyond. Nature Reviews Genetics, 6(8), 611–622. [CrossRef]
- Bilban M, Haschemi A, Wegiel B, Chin BY, Wagner O, Otterbein LE. Heme oxygenase and carbon monoxide initiate homeostatic signaling. J Mol Med (Berl). 2008;86(3):267-279. [CrossRef]
- Boccitto, M. , & Wolin, S. L. (2019). Ro60 and Y RNAs: structure, functions, and roles in autoimmunity. Critical reviews in biochemistry and molecular biology, 54(2), 133–152. [CrossRef]
- Bochman, M. L. , & Schwacha, A. (2009). The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiology and molecular biology reviews : MMBR, 73(4), 652–683. [CrossRef]
- Bolt and Their, 2006: Bolt, H. M., & Their, R. (2006). Relevance of the deletion polymorphisms of the glutathione S-transferases GSTT1 and GSTM1 in pharmacology and toxicology. Current Drug Metabolism, 7(6), 613-628. [CrossRef]
- Borchert et al., 2018: Borchert, A., Kalms, J., Roth, S., Rademann, J., & Brückner, M. (2018). Glutathione peroxidase 4: A key to ferroptosis and a potential therapeutic target to counteract chemoresistance. Cell Chemical Biology, 25(4), 482-493.e3. [CrossRef]
- Boutilier RG, St-Pierre J. Surviving hypoxia without really dying. Comp Biochem Physiol A Mol Integr Physiol. 2000 Sep;126(3):481-90.
- Borisov VB, Forte E, Giuffrè A, et al. Carbon Monoxide in Biology and Medicine: A Guide for Clinicians and Researchers. Biophys Rev. 2021;13(4):703-709.
- Brazda V, Kolomaznik J, Lysek J, Bartas M, Fojta M, Stastny J, et al. G4Hunter web application: a web server for G-quadruplex prediction. Bioinformatics. 2019;35(17):3493-3495.
- Cadet J, Davies KJA. Oxidative DNA damage & repair: An introduction. Free Radic Biol Med. 2017;107:2-12. [CrossRef]
- Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev. 2003;83(4):1153-1181. [CrossRef]
- Chang, Y. F. , et al. (2007). Nonsense-mediated mRNA decay: A mechanistic perspective. Nature Reviews Molecular Cell Biology, 8(7), 593–606. [CrossRef]
- Che, W. , et al. (1997). Selective induction of heparin-binding epidermal growth factor-like growth factor by methylglyoxal and 3-deoxyglucosone in rat aortic smooth muscle cells. The Journal of Biological Chemistry, 272(28), 18453–18459. [CrossRef]
- Chen, S. , & Pandolfi, P. P. (2016). RNA editing and the epitranscriptome in cancer. Cancer Discovery, 6(6), 565-571. [CrossRef]
- Conde-Pérezprina, J. C. , Luna-López, A., González-Puertos, V. Y., Zenteno-Savín, T., León-Galván, M. A., & Königsberg, M. (2012). DNA MMR systems, microsatellite instability and antioxidant activity variations in two species of wild bats: Myotis velifer and Desmodus rotundus, as possible factors associated with longevity. Age (Dordrecht, Netherlands), 34(6), 1473–1492. [CrossRef]
- Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21(2):121-131. [CrossRef]
- Cox, D. B. T. , et al. (2017). RNA editing with CRISPR-Cas13. Science, 358(6366), 1019–1027. [CrossRef]
- Cooke, M. S. , et al. (2003). Oxidative DNA damage: mechanisms, mutation, and disease. FASEB Journal, 17(10), 1195-1214. [CrossRef]
- Cui L, Ma R, Cai J, et al. RNA modifications: importance in immune cell biology and related diseases. Signal Transduct Target Ther. 2022;7(1):334. Published 2022 Sep 22. [CrossRef]
- Cech T., R. (2018). A Lifelong Passion for All Things Ribonucleic. Cell, 175(1), 14–17. [CrossRef]
- D'Annibale V, Nardi AN, Amadei A, D'Abramo M. Theoretical Characterization of the Reduction Potentials of Nucleic Acids in Solution. J Chem Theory Comput. 2021;17(3):1301-1307. [CrossRef]
- Defenouillère, Q. , et al. (2013). Rqc1 and Ltn1 prevent accumulation of peptidyl-tRNA on stalled ribosomes. Nature Structural & Molecular Biology, 20(6), 659–666. [CrossRef]
- Degtyareva NP, Chen L, Mieczkowski P, Petes TD, Doetsch PW (2008) Chronic oxidative DNA damage due to DNA repair defects causes chromosomal instability in Saccharomyces cerevisiae. Mol Cell Biol 28: 5432–5445.
- Doma, M. K. , & Parker, R. (2006). RNA quality control in eukaryotes: Nonsense-mediated decay and related pathways. Nature, 440(7083), 561–567. [CrossRef]
- Dominissini, D. , et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature, 485(7397), 201–206. [CrossRef]
- Dugbartey, G. J. (2024). Cellular and molecular mechanisms of cell damage and cell death in ischemia–reperfusion injury. Molecular Biology Reports, 51(2), 2348–2360. [CrossRef]
- Emre Y, Nübel T. Uncoupling protein UCP2: when mitochondrial activity meets immunity. FEBS Lett. 2010;584(8):1437-1442. [CrossRef]
- Esteller, M. (2011). Non-coding RNAs in human disease. Nature Reviews Genetics, 12(12), 861–874. [CrossRef]
- Evert BA, Salmon TB, Song B, Jingjing L, Siede W, et al. (2004) Spontaneous DNA damage in Saccharomyces cerevisiae elicits phenotypic properties similar to cancer cells. J Biol Chem 279: 22585–22594.
- Fischer, L. R. , et al. (2011). SOD1 deficiency and neurodegeneration. Experimental Neurology, 233(1), 89–97. [CrossRef]
- Fritsch EF, Ott PA. Personalized Cancer Vaccines Directed against Tumor Mutations: Building Evidence from Mice to Humans. Cancer Res. 2024;84(7):953-955. [CrossRef]
- Gastelum S, Michael AF, Bolger TA. Saccharomyces cerevisiae as a research tool for RNA-mediated human disease. Wiley Interdiscip Rev RNA. Published online September 6, 2023. [CrossRef]
- Gámez-Valero A, Guisado-Corcoll A, Herrero-Lorenzo M, Solaguren-Beascoa M, Martí E. Non-Coding RNAs as Sensors of Oxidative Stress in Neurodegenerative Diseases. Antioxidants (Basel). 2020 Nov 8;9(11):1095. [CrossRef]
- Giani M, Pire C, Martínez-Espinosa RM. Bacterioruberin: Biosynthesis, Antioxidant Activity, and Therapeutic Applications in Cancer and Immune Pathologies. Mar Drugs. 2024;22(4):167. Published 2024 Apr 9. [CrossRef]
- Goodchild, A. , et al. (2009). Sequence determinants of innate immune activation by short interfering RNAs. BMC Immunology, 10(40). https://bmcimmunol.biomedcentral.com/articles/10.1186/1471-2172-10-40.
- Gordon, A. J. E. , et al. (2020). RNA polymerase errors and their implications for transcript fidelity. Trends in Genetics, 36(3), 153–167. [CrossRef]
- Greider, C. W. , & Blackburn, E. H. (1985). Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell, 43(2 Pt 1), 405-413. [CrossRef]
- Greider, C. W. (1996). Telomere length regulation. Annual Review of Biochemistry, 65, 337–365. [CrossRef]
- Griko, Y, Loftus, D., and Stolc, V. Metabolic Suppression: A Promising Solution to Unlock the Future of Space Travel. Journal of Tourism and Hospitality 12.6 (2024).
- Hahm JY, Park J, Jang ES, Chi SW. 8-Oxoguanine: from oxidative damage to epigenetic and epitranscriptional modification. Exp Mol Med. 2022;54(10):1626-1642. [CrossRef]
- Hainsworth AH, Drinkhill MJ. Therapeutic potential of hypometabolism for protection against ischemia-reperfusion injury. Pharmacol Ther. 2023 Jul;247:108461.
- Halliwell, B, Adhikary, A, Dingfelder, M, Dizdaroglu, M. Hydroxyl radical is a significant player in oxidative DNA damage in vivo. Chem Soc Rev. 2021;50(15):8355-8360. [CrossRef]
- Han, L. , et al. (2015). The genomic landscape and clinical relevance of RNA editing in cancer. Nature Communications, 6, 6745. [CrossRef]
- Hashimoto S, Noguchi E, Bando H, et al. Neoantigen prediction in human breast cancer using RNA sequencing data. Cancer Sci. 2021;112(1):465-475. [CrossRef]
- Hemagirri M, Sasidharan S. Biology of aging: Oxidative stress and RNA oxidation. Mol Biol Rep. 2022;49(6):5089-5105. [CrossRef]
- Henrich, L. , Kiessling, I., Steimer, M. et al. Circadian dependency of microglial heme oxygenase-1 expression and inflammation determine neuronal injury in hemorrhagic stroke. J Inflamm 20, 43 (2023). [CrossRef]
- Hermes-Lima M, Storey JM, Storey KB. Antioxidant defenses and metabolic depression. The hypothesis of preparation for oxidative stress in land snails. Comp Biochem Physiol B Biochem Mol Biol. 1998;120(3):437-448. [CrossRef]
- Higuchi, M. , et al. (2000). RNA editing of AMPA receptor subunit GluR-B: A molecular mechanism for Q/R site-selective and stage-specific RNA processing in the brain. Nature, 406, 78–81. [CrossRef]
- Houseley, J. , & Tollervey, D. (2009). The many pathways of RNA degradation. Cell, 136(4), 763–776. [CrossRef] [PubMed]
- Iraqui I, Kienda G, Soeur J, Faye G, Baldacci G, et al. (2009) Peroxiredoxin Tsa1 is the key peroxidase suppressing genome instability and protecting against cell death in Saccharomyces cerevisiae. PLoS Genet 5(6): e1000524.
- Iqbal MJ, Kabeer A, Abbas Z, et al. Interplay of oxidative stress, cellular communication and signaling pathways in cancer. Cell Commun Signal. 2024;22(1):7. Published 2024 Jan 2. [CrossRef]
- Jarrous N, Liu F. Human RNase P: overview of a ribonuclease of interrelated molecular networks and gene-targeting systems. RNA. 2023;29(3):300-307. [CrossRef]
- Jiang J, Chan A, Ali S, et al. Hydrogen Sulfide--Mechanisms of Toxicity and Development of an Antidote. Sci Rep. 2016;6:20831. Published 2016 Feb 15. [CrossRef]
- Jiang, M. , Wang, H., Liu, Z., Lin, L., Wang, L., Xie, M., Li, D., Zhang, J., & Zhang, R. (2020). Endoplasmic reticulum stress-dependent activation of iNOS/NO-NF-κB signaling and NLRP3 inflammasome contributes to endothelial inflammation and apoptosis associated with microgravity. FASEB journal : official publication of the Federation of American Societies for Experimental Biology, 34(8), 10835–10849. [CrossRef]
- Jiang, T. , Shi, T., Zhang, H., Hu, J., Song, Y., Wei, J., Ren, S., & Zhou, C. (2019). Tumor neoantigens: from basic research to clinical applications. Journal of hematology & oncology, 12(1), 93. [CrossRef]
- Johnson RJ, Andrews P, Benner SA, Oliver W. Theodore E. Woodward award. The evolution of obesity: insights from the mid-Miocene [published correction appears in Trans Am Clin Climatol Assoc. 2013;124:294. Trans Am Clin Climatol Assoc. 2010;121:295-308.
- Jorgensen, A, Brandslund, I, Ellervik, C, et al. Oxidative Stress-Induced Damage to RNA and DNA and Mortality in Individuals with Psychiatric Illness. JAMA Psychiatry. 2024;81(5):516-520. [CrossRef]
- Jorgensen, A, Brandslund, I, Ellervik, C, et al. Specific prediction of mortality by oxidative stress-induced damage to RNA vs. DNA in humans. Aging Cell. 2023;22(6):e13839. [CrossRef]
- Juan CA, Pérez de la Lastra JM, Plou FJ, Pérez-Lebeña E. The Chemistry of Reactive Oxygen Species (ROS) Revisited: Outlining Their Role in Biological Macromolecules (DNA, Lipids and Proteins) and Induced Pathologies. Int J Mol Sci. 2021;22(9):4642. Published 2021 Apr 28. [CrossRef]
- Lamarck, J.-B. (1809). Philosophie zoologique, ou exposition des considérations relatives à l'histoire naturelle des animaux; à la diversité de leur organisation et des 1 facultés qu'ils en obtiennent; aux causes physiques qui maintiennent en eux la vie et donnent lieu aux mouvemens qu'ils exécutent; enfin, à celles qui produisent, 2 les unes le sentiment et les autres l'intelligence. 3 Paris: Dentu.
- Li M, Wang IX, Li Y, et al. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333(6038):53-58. [CrossRef]
- Li W, Zhang C, Jackson K, et al. UCP2 knockout suppresses mouse skin carcinogenesis. Cancer Prev Res (Phila). 2015;8(6):487-491. [CrossRef]
- Lo et al., 2007: Lo, H. W., Ali-Osman, F., & Xia, W. (2007). Genetic polymorphism and function of glutathione S-transferases in tumor drug resistance. Current Opinion in Pharmacology, 7(4), 367-374. [CrossRef]
- Kiani, L. Genetic protection against Alzheimer disease. Nat Rev Neurol 20, 316 (2024). [CrossRef]
- Kjær LK, Cejvanovic V, Henriksen T, et al. Cardiovascular and All-Cause Mortality Risk Associated With Urinary Excretion of 8-oxoGuo, a Biomarker for RNA Oxidation, in Patients With Type 2 Diabetes: A Prospective Cohort Study. Diabetes Care. 2017;40(12):1771-1778. [CrossRef]
- Kong Q, Lin CL. Oxidative damage to RNA: mechanisms, consequences, and diseases. Cell Mol Life Sci. 2010;67(11):1817-1829. [CrossRef]
- Korczowska-Łącka I, Słowikowski B, Piekut T, Hurła M, Banaszek N, Szymanowicz O, Jagodziński PP, Kozubski W, Permoda-Pachuta A, Dorszewska J. Disorders of Endogenous and Exogenous Antioxidants in Neurological Diseases. Antioxidants (Basel). 2023 Sep 29;12(10):1811. [CrossRef]
- Kumar, A. , & Atkinson, G. (2023). GlyNAC supplementation improves age-related defects in older humans: A randomized clinical trial. Journal of Gerontology: Biological Sciences, 78(1), 75–83. [CrossRef]
- Kumar D, Viberg J, Nilsson AK, Chabes A (2010) Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res 38: 3975–3983.
- Kumar D, Abdulovic AL, Viberg J, Nilsson AK, Kunkel TA, et al. (2011) Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res 39: 1360–1371.
- Ma S, Han C, Wang D, Xie Z, Liu J, Li Z. Synthetic torpor: a paradigm shift in clinical practice? Front Pharmacol. 2022 Oct 28;13:1041355.
- MacNee, W. (2001). Oxidative stress and lung inflammation in airways disease. European Journal of Pharmacology, 429(1-3), 195–207.
- Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9(4):e1003468. [CrossRef]
- Matute JD, Arias AA, Wright NA, et al. A new genetic subgroup of chronic granulomatous disease with autosomal recessive mutations in p40 phox and selective defects in neutrophil NADPH oxidase activity. Blood. 2009;114(15):3309-3315. [CrossRef]
- Meier, J. C. , et al. (2020). RNA editing and beyond: The epitranscriptomic machinery as a key regulator of cell physiology and pathology. Frontiers in Molecular Neuroscience, 13, 566864. [CrossRef]
- Meyer, K. D. , et al. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell, 149(7), 1635–1646. [CrossRef]
- Mihaljevic, O. , et al. (2020). Oxidative stress and DNA damage in critically ill patients with sepsis. Critical Care, 24(1), 414.
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017;22(1):11-19. [CrossRef]
- Munteanu C, Turnea MA, Rotariu M. Hydrogen Sulfide: An Emerging Regulator of Oxidative Stress and Cellular Homeostasis-A Comprehensive One-Year Review. Antioxidants (Basel). 2023;12(9):1737. Published 2023 Sep 7. [CrossRef]
- Murray DB, Lloyd D. Multiple Rediscoveries and Misconceptions; the Yeast Metabolic Oscillation. Function (Oxf). 2021;2(5):zqab039. Published 2021 Sep 3. [CrossRef]
- Ng, PC, Hendry-Hofer, TB, Witeof,AE, et al. Hydrogen Sulfide Toxicity: Mechanism of Action, Clinical Presentation, and Countermeasure Development. J Med Toxicol. 2019;15(4):287-294. [CrossRef]
- Nislow C, Lee AY, Allen PL, et al. Genes required for survival in microgravity revealed by genome-wide yeast deletion collections cultured during spaceflight. Biomed Res Int. 2015;2015:976458. [CrossRef]
- Nunomura, A. , et al. (1999). RNA oxidation is a prominent feature of vulnerable neurons in Alzheimer’s disease. Journal of Neuroscience Research, 56(6), 724–728.
- Navaratnam N, Bhattacharya S, Fujino T, Patel D, Jarmuz AL, Scott J. Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell. 1995;81(2):187-195. [CrossRef]
- O'Brien E, Holt ME, Thompson MK, et al. The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science. 2017;355(6327):eaag1789. [CrossRef]
- Pleskova SN, Erofeev AS, Vaneev AN, et al. ROS Production by a Single Neutrophil Cell and Neutrophil Population upon Bacterial Stimulation. Biomedicines. 2023;11(5):1361. Published 2023 May 4. [CrossRef]
- Panda, S. , Chatterjee, O., Mukherjee, G., Chatterjee, S. (2023). Human Diseases Induced by Oxidative Damage in DNA. In: Chatterjee, S., Chattopadhyay, S. (eds) Nucleic Acid Biology and its Application in Human Diseases. Springer, Singapore.
- Panatta E, Zampieri C, Melino G, Amelio I. Understanding p53 tumour suppressor network. Biol Direct. [CrossRef]
- Podlutsky, A. J. , Khritankov, A. M., Ovodov, N. D., & Austad, S. N. (2005). A new field record for bat longevity. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences, 60(11), 1366–1368.
- Pang Z, Lu MM, Zhang Y, et al. Neoantigen-targeted TCR-engineered T cell immunotherapy: current advances and challenges. Biomark Res. 2023;11(1):104. Published 2023 Dec 1. [CrossRef]
- Paz-Yaacov, N. , Bazak, L., Buchumenski, I., et al. (2015). Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Reports, 13(1), 267–276. [CrossRef] [PubMed]
- Peng G, Tang Z, Xiang Y, Chen W. Glutathione peroxidase 4 maintains a stemness phenotype, oxidative homeostasis and regulates biological processes in Panc-1 cancer stem-like cells. Oncol Rep. 2019;41(2):1264-1274. [CrossRef]
- Peng M, Mo Y, Wang Y, et al. Neoantigen vaccine: an emerging tumor immunotherapy. Mol Cancer. 2019;18(1):128. Published 2019 Aug 23. [CrossRef]
- Poetsch A., R. (2020). The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Computational and structural biotechnology journal, 18, 207–219. [CrossRef]
- Puga I, Cols M, Barra CM, et al. B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen [published correction appears in Nat Immunol. 2014 Feb;15(2):205]. Nat Immunol. 2011;13(2):170-180. Published 2011 Dec 25. [CrossRef]
- Ragu S, Faye G, Iraqui I, Masurel-Heneman A, Kolodner RD, et al. (2007) Oxygen metabolism and reactive oxygen species cause chromosomal rearrangements and cell death. Proc Natl Acad Sci U S A 104: 9747–9752.
- Rahman I, Adcock IM. Oxidative stress and redox regulation of lung inflammation in COPD. Eur Respir J. 2006;28(1):219-242. [CrossRef]
- Roundtree, I. A. , Evans, M. E., Pan, T., & He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell, 169(7), 1187–1200. [CrossRef] [PubMed]
- Samanta, M. P. , Tongprasit, W., Sethi, H., Chin, C. S., & Stolc, V. (2006). Global identification of noncoding RNAs in Saccharomyces cerevisiae by modulating an essential RNA processing pathway. Proceedings of the National Academy of Sciences of the United States of America, 103(11), 4192–4197. [CrossRef] [PubMed]
- Saxena P, Selvaraj K, Khare SK, Chaudhary N. Superoxide dismutase as multipotent therapeutic antioxidant enzyme: Role in human diseases. Biotechnol Lett. 2022;44(1):1-22. [CrossRef]
- Seo, Y. S. , & Kang, Y. H. (2018). The Human Replicative Helicase, the CMG Complex, as a Target for Anti-cancer Therapy. Frontiers in molecular biosciences, 5, 26. [CrossRef]
- Shay, J. W. , & Wright, W. E. (2019). Telomeres and telomerase: Three decades of progress. Nature Reviews Genetics, 20(5), 299–309. [CrossRef]
- Schieber, M. , & Chandel, N. S. (2014). ROS function in redox signaling and oxidative stress. Current Biology, 24(10), R453–R462.
- Simms CL, Zaher HS. Quality control of chemically damaged RNA. Cell Mol Life Sci. 2016;73(19):3639-3653. [CrossRef]
- Singh et al., 2008: Singh, M., Khan, A. J., & Singh, K. (2008). Association of polymorphism in superoxide dismutase (SOD2) gene with Parkinson's disease in North Indian population. Indian Journal of Biochemistry & Biophysics, 45(5), 337-342. https://www.ncbi.nlm.nih.gov/pubmed/19179754. 1917.
- Slavov N, Botstein D. Coupling among growth rate response, metabolic cycle, and cell division cycle in yeast. Mol Biol Cell. 2011;22(12):1997-2009. [CrossRef]
- Sohn HY, Murray DB, Kuriyama H. Ultradian oscillation of Saccharomyces cerevisiae during aerobic continuous culture: hydrogen sulphide mediates population synchrony. Yeast. 2000;16(13):1185-1190. [CrossRef]
- Sontz PA, Mui TP, Fuss JO, Tainer JA, Barton JK. DNA charge transport as a first step in coordinating the detection of lesions by repair proteins. Proc Natl Acad Sci U S A. 2012;109(6):1856-1861. [CrossRef]
- Sorrentino ZA, Vijayaraghavan N, Gorion KM, et al. Physiological C-terminal truncation of α-synuclein potentiates the prion-like formation of pathological inclusions. J Biol Chem. 2018;293(49):18914-18932. [CrossRef]
- Springer Nature Switzerland AG. (2023). RNA Structure and Function (Vol. 14). Springer Nature.
- Stolc, V. Genetic control of blood neutrophil concentration in the rat. J Immunogenet. 1988;15(5-6):345-351. [CrossRef]
- Stolc V, Altman S. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes Dev. 1997;11(18):2414-2425. [CrossRef]
- Stolc V, Shmygelska A, Griko Y. Adaptation of organisms by resonance of RNA transcription with the cellular redox cycle. PLoS One. 2011;6(9):e25270. [CrossRef]
- Stolc, V, Karhanek M, Freund F, Griko Y, Loftus DJ, Ohayon MM. Metabolic stress in space: ROS-induced mutations in mice hint at a new path to cancer. Redox Biol. Published online October 16, 2024. [CrossRef]
- Storey KB, Storey JM. Metabolic rate depression in animals: transcriptional and translational controls. Biol Rev Camb Philos Soc. 2004;79(1):207-233. [CrossRef]
- Szade, A. , Szade, K., Mahdi, M. et al. The role of heme oxygenase-1 in hematopoietic system and its microenvironment. Cell. Mol. Life Sci. 78, 4639–4651 (2021). [CrossRef]
- Tanaka, M. , et al. (2007). Oxidative RNA damage and lifespan in yeast. Molecular Biology of the Cell, 18(11), 4647–4655.
- Tasaki, E. , Sakurai, H., Nitao, M., Matsuura, K., & Iuchi, Y. (2017). Uric acid, an important antioxidant contributing to survival in termites. PloS one, 12(6), e0179426. [CrossRef] [PubMed]
- Taylor, KE, Miller LG, Contreras LM. RNA-binding proteins that preferentially interact with 8-oxoG-modified RNAs: our current understanding. Biochem Soc Trans. 2024;52(1):111-122. [CrossRef]
- Tesi N, van der Lee S, Hulsman M, et al. Cognitively healthy centenarians are genetically protected against Alzheimer's disease. Alzheimers Dement. 2024;20(6):3864-3875. [CrossRef]
- Thalau P, Ritz T, Stapput K, Wiltschko R, Wiltschko W. Magnetic compass orientation of migratory birds in the presence of a 1.315 MHz oscillating field. Naturwissenschaften. 2005;92(2):86-90. [CrossRef]
- Thomas EN, Simms CL, Keedy HE, Zaher HS. Insights into the base-pairing preferences of 8-oxoguanosine on the ribosome. Nucleic Acids Res. 2019;47(18):9857-9870. [CrossRef]
- Tian L, Luo Y, Ren J, Zhao C. The Role of Oxidative Stress in Hypomagnetic Field Effects. Antioxidants (Basel). 2024;13(8):1017. Published 2024 Aug 21. [CrossRef]
- Time. "A Melanoma Vaccine Showed Promising Results in a New Study." TIME, 17 Apr. 2023, https://time.com/6984417/melanoma-vaccine-moderna-mrna/.
- Tonegawa, S. Somatic generation of antibody diversity. Nature. 1983;302(5909):575-581. [CrossRef]
- Toung, J. M. , et al. (2014). RNA editing as a source of tumor-specific antigenic diversity in immunotherapy. PLoS ONE, 9(11), e112040.
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes [published correction appears in Science. 2006 Feb 17;311(5763):954]. Science. 2005;310(5751):1152-1158. [CrossRef]
- Tu BP, McKnight SL. Evidence of carbon monoxide-mediated phase advancement of the yeast metabolic cycle. Proc Natl Acad Sci U S A. 2009;106(34):14293-14296. [CrossRef]
- Turrens, JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552(Pt 2):335-344. [CrossRef]
- Vaghf, A, Khansarinejad B, Ghaznavi-Rad E, Mondanizadeh M. The role of microRNAs in diseases and related signaling pathways. Mol Biol Rep. 2022;49(7):6789-6801. [CrossRef]
- Vauclare P, Wulffelé J, Lacroix F, et al. Stress-induced nucleoid remodeling in Deinococcus radiodurans is associated with major changes in Heat Unstable (HU) protein dynamics. Nucleic Acids Res. 2024;52(11):6406-6423. [CrossRef]
- Varshney D, Spiegel J, Zyner K, Tannahill D, Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat Rev Mol Cell Biol. 2020;21(8):459-474. [CrossRef]
- von Zglinicki, T. (2002). Oxidative stress shortens telomeres. Trends in Biochemical Sciences, 27(7), 339–344. [CrossRef]
- Wang, I. X. , Grunseich, C., Chung, Y. G., Kwak, H., Ramrattan, G., Zhu, Z., & Cheung, V. G. 2016. RNA-DNA sequence differences in Saccharomyces cerevisiae. Genome research, 26: 1544–1554.
- Wang et al., 2016: Wang, M., Li, Y., Lin, L., Song, G., & Deng, T. (2016). GSTM1 null genotype and GSTP1 Ile105Val polymorphism are associated with Alzheimer's disease: A meta-analysis. Molecular Neurobiology, 53(2), 1355-1364. [CrossRef]
- Wang, M. , et al. (2021). Systematic understanding of pathophysiological mechanisms of oxidative stress-related conditions—diabetes mellitus, cardiovascular diseases, and ischemia–reperfusion injury. Frontiers in Cardiovascular Medicine, 8, 649785. [CrossRef]
- Wheeler, HB, Madrigal, AA, Chaim, IA. Mapping the future of oxidative RNA damage in neurodegeneration: Rethinking the status quo with new tools. Proc Natl Acad Sci U S A. 2024;121(46):e2317860121. [CrossRef]
- Wu F, Du H, Overbey E, et al. Single-cell analysis identifies conserved features of immune dysfunction in simulated microgravity and spaceflight. Nat Commun. 2024;15(1):4795. Published 2024 Jun 11. [CrossRef]
- Wu S, Jiang L, Lei L, et al. Crosstalk between G-quadruplex and ROS. Cell Death Dis. 2023;14(1):37. [CrossRef]
- Wurtmann, E. J. , & Wolin, S. L. (2009). RNA under attack: Cellular handling of RNA damage. Critical Reviews in Biochemistry and Molecular Biology, 44(2-3), 139-149. [CrossRef]
- Wu Z, Lou Y, Jin W, et al. Relationship of the p22phox (CYBA) gene polymorphism C242T with risk of coronary artery disease: a meta-analysis [published correction appears in PLoS One. 2013;8(12). doi:10.1371/annotation/7f489b96-e43f-4c6d-86d7-0689c8a55eba]. PLoS One. 2013;8(9):e70885. Published 2013 Sep 5. [CrossRef]
- Xu, L, Wang, W, Chong, J, Shin, JH, Xu, J, Wang, D. RNA polymerase II transcriptional fidelity control and its functional interplay with DNA modifications. Crit Rev Biochem Mol Biol. 2015;50(6):503-19. [CrossRef]
- Yamamura Y, Kawamura Y, Oka K, Miura K. Carcinogenesis resistance in the longest-lived rodent, the naked mole-rat. Cancer Sci. 2022;113(12):4030-4036. [CrossRef]
- Yan LL, Simms CL, McLoughlin F, Vierstra RD, Zaher HS. Oxidation and alkylation stresses activate ribosome-quality control. Nat Commun. 2019;10(1):5611. Published 2019 Dec 9. [CrossRef]
- Yang, S. , et al. (2024). MiR-574-5p activates human TLR8 to promote autoimmune signaling and inflammation. Cell Communication and Signaling, 22(220). https://biosignaling.biomedcentral.com/articles/10.1186/s12964-024-01601-1.
- Yuan, J, Xu, L, Bao, HJ, Wang, JL, Zhao, Y, Chen, S. Biological roles of A-to-I editing: implications in innate immunity, cell death, and cancer immunotherapy. J Exp Clin Cancer Res. 2023;42(1):149. Published 2023 Jun 17. [CrossRef]
- Zhou, C. , Wei, Z., Zhang, L., Yang, Z., & Liu, Q. (2020). Systematically Characterizing A-to-I RNA Editing Neoantigens in Cancer. Frontiers in oncology, 10, 593989. [CrossRef]
- Zhou, X. , Li, Y., & Yan, Y. (2011). RNA epigenetics and RNA-protein interactions: Connections with DNA methylation. Current Genomics, 12(8), 604–612. [CrossRef]

 |
 |
 |
 |
 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





