1. Introduction
The cereal crop foxtail millet (
Setaria italica) holds considerable significance, originating from the long-term domestication of green foxtail (
Setaria viridis) and possessing a rich historical and cultural background [
1]. Rapid population growth poses significant challenges to food and nutritional security worldwide [
2], and foxtail millet emerges as a potential candidate to address these issues [
3]. Its drought tolerance and adaptability make it a widely cultivated crop in many developing countries across Asia and Africa. Abiotic stresses reduce millet yields in regions with low rainfall, significantly impacting cultivation productivity and financial returns [
4]. Under such conditions, the yield of millet can decrease substantially, leading to not only direct economic losses but also potential food security and livelihood challenges for rural populations. Therefore, improving the adaptability and yield of millet in harsh environments is essential for ensuring food security and promoting sustainable agricultural development.
Transcription factors in plants play a vital role in controlling gene expression [
5]. These factors serve as essential regulators in various plant processes, such as flowering [
6,
7], disease resistance [
8], circadian rhythms [
9], cell differentiation [
10], hormonal responses [
11], and the biotransformation of carbon and nitrogen [
12]. The BRX gene family is a widely distributed group of plant genes, initially identified in studies on root development and morphological regulation. Advancements in molecular biology have resulted in the identification and characterization of an increasing count of BRX family members. These genes typically possess conserved structural features, including multiple functional domains such as the BRX domain and other potential regulatory regions [
13]. They exhibit high sequence similarity across various plant species. Investigations have shown that BRX gene family members influence the development of multiple organs, including roots, stems, and leaves, by modulating plant hormone signaling pathways, particularly auxin [
14,
15]. Additionally, the function of BRX gene members in responding to environmental stimuli and abiotic stresses is increasingly recognized.
The BRX gene family is crucial to root development, morphological regulation, and environmental stress response of many crops. BRX gene expression promotes root growth and branching in wheat and barley, enhancing the plants’ ability to absorb water and nutrients, thereby improving stress tolerance and yield stability. This effect is particularly pronounced under drought conditions, enabling these crops to maintain robust growth in adverse environments [
16,
17]. BRX gene family members are involved in not only root development but also stem growth and tillering regulation in rice, contributing to higher photosynthetic efficiency and yield during the growing season [
18]. BRX gene family members are crucial to stress response in soybeans, with studies indicating that they enhance adaptability to unfavorable conditions such as drought and salinity by regulating signaling pathways, ultimately leading to improved yield and biomass production [
19]. Additionally, the BRX gene family is fundamental to vegetable crops such as Chinese cabbage; it is involved in fruit development and maturation, influencing fruit size, shape, and nutritional content. BRX gene family members are strongly linked to the bolting trait in Chinese cabbage, highlighting their significance in not only staple crops but also high-value vegetable varieties [
20]. In summary, the BRX gene family plays diverse roles across different crops, contributing to plant growth, development, yield enhancement, and environmental adaptability, providing an essential genetic foundation for sustainable agriculture.
This study identified 15 BRX gene family members in both green foxtail and foxtail millet, analyzing their conserved protein motifs, phylogenetic relationships, locations, and cis-acting regulatory elements in promoters. Based on these analyses, the expression profiles of this gene family were tested in five parts of foxtail millet and under different stress conditions. The findings suggested a significant role of the BRX gene family in stress responses in foxtail millet. This study offers important resources for future studies on the functional roles and molecular mechanisms of the BRX gene family, laying the groundwork for breeding stress-resistant crop varieties.
2. Results
2.1. Identification of the BRX Gene Family in Foxtail Millet and Green Foxtail
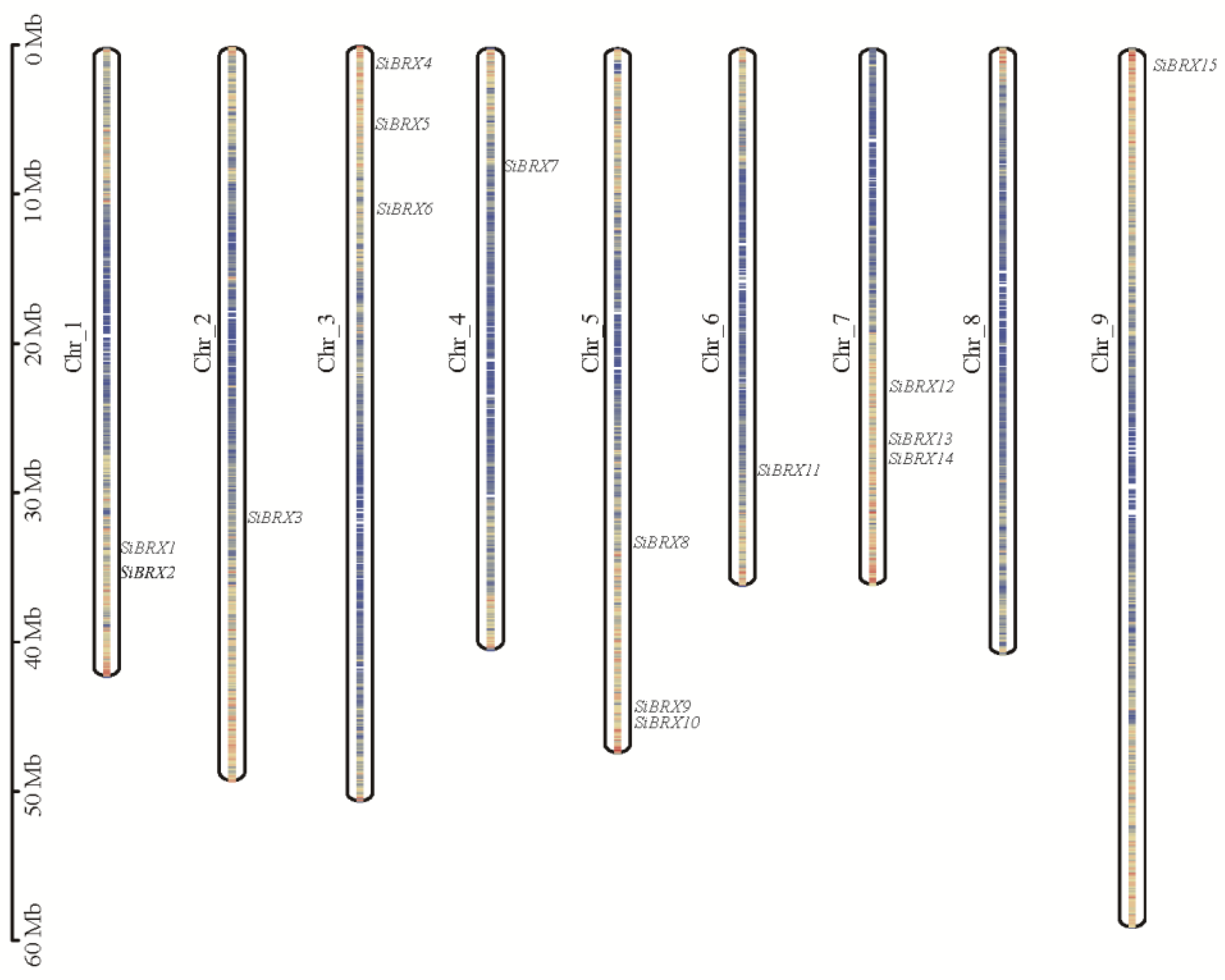
A total of 15 BRX genes were identified (
Table 1) in both green foxtail and foxtail millet, named
SiBRX1 to
SiBRX15 and
SvBRX1 to
SvBRX15, respectively, based on their chromosomal locations. As shown in
Figure 1, the 15
SiBRX genes were distributed across 8 chromosomes of foxtail millet. Chromosomes 3, 5, and 7 harbored the highest number of BRX gene family members, with three genes each.
SiBRX9 and
SiBRX10 on chromosome 5, and
SiBRX13 and
SiBRX14 on chromosome 7, were clustered. In contrast, chromosomes 2, 4, 6, and 9 each contained only one SiBRX gene, chromosome 1 had two SiBRX genes, and chromosome 8 lacked any detectable SiBRX genes.
Further examination of the fundamental physicochemical characteristics and subcellular localization of SiBRX proteins revealed that their amino acid sequences ranged from 300 to 1092 residues, with molecular weights between 115.18 and 1116.49 kDa. The isoelectric points (pI) of SiBRX proteins ranged from 4.91 to 9.18, with the majority having a pI greater than 7.0, indicating a higher proportion of basic amino acids. The instability index analysis showed that SiBRX1 had the lowest instability index of 37.32, whereas SiBRX15 had the highest value at 55.64. SiBRX1 and SiBRX8 were stable proteins, whereas the remaining SiBRX proteins exhibited instability (instability index > 40). The average hydropathy index of all SiBRX proteins was less than 0, suggesting that these proteins were hydrophilic. The aliphatic index of SiBRX proteins ranged from 48.3 to 78.42. Subcellular localization predictions indicated that eight SiBRX proteins were localized to the extracellular membrane whereas the remaining seven were located in the extracellular space. These findings provide crucial genetic and biochemical insights into the functions of the SiBRX gene family and their roles in the growth and development of foxtail millet.
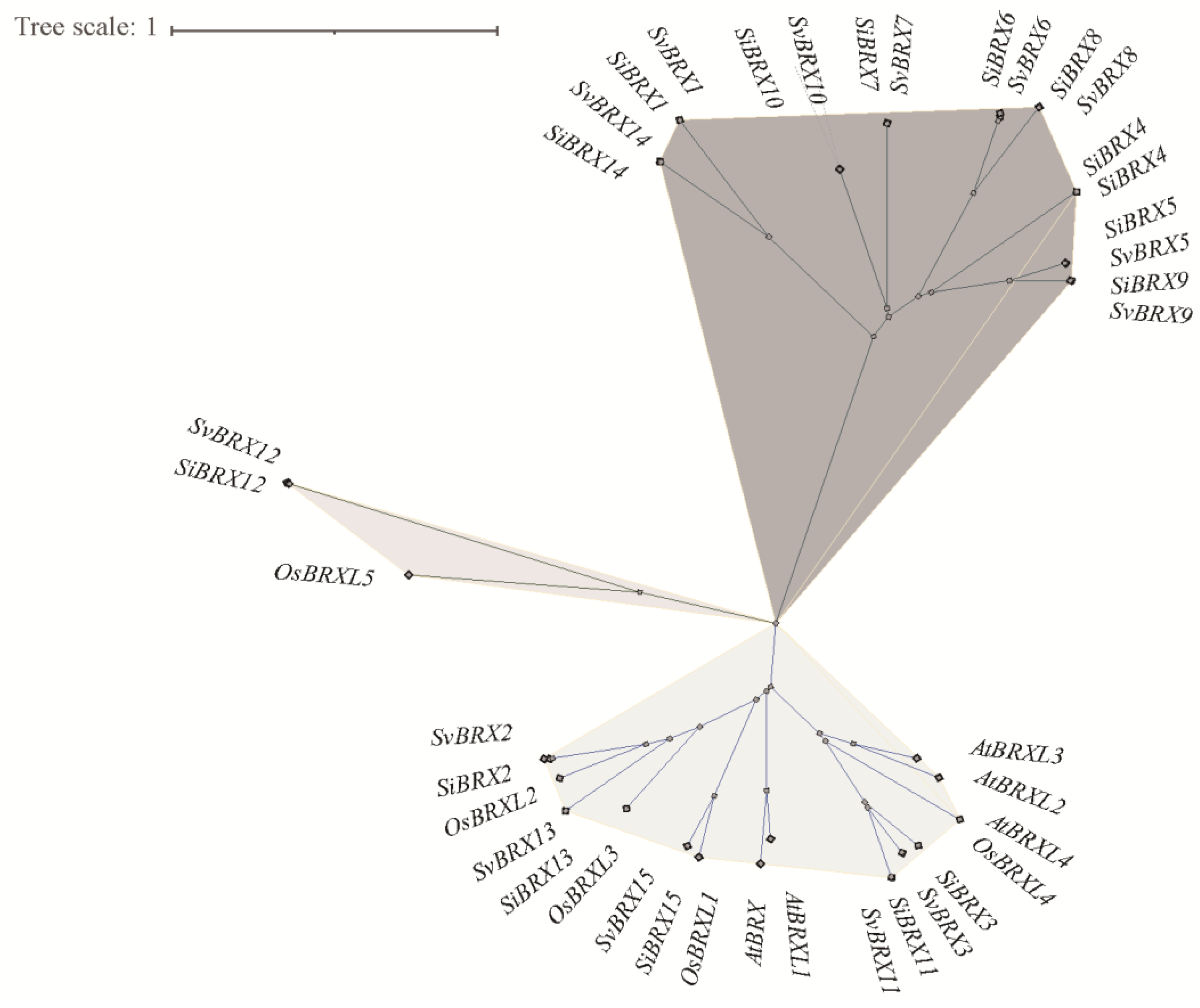
2.2. Phylogenetic Analysis of the BRX Gene Family
All This study used 30 BRX protein sequences from green foxtail and foxtail millet, along with 10 BRX genes from rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana), to construct a phylogenetic tree using the NJ method based on multiple sequence alignment analysis (Figure. 2). This analysis aimed to elucidate the phylogenetic relationships among BRX family genes. The 30 BRX proteins were clustered into 3 groups through 1000 bootstrap resampling iterations: Group I, Group II, and Group III. Group I included 18 genes, 9 from foxtail millet (SiBRXs) and 9 from green foxtail (SvBRXs). This group did not contain any BRX genes from Arabidopsis or rice. Group II consisted of 19 genes, including all BRX genes from Arabidopsis, as well as 5 SiBRXs and 5 SvBRXs. Group III, the smallest group, contained only three members: SvBRX12, SiBRX12, and OsBRXL5. Further phylogenetic analysis revealed that BRX genes in green foxtail and foxtail millet exhibited a one-to-one correspondence and shared similar genetic distances, suggesting that these genes in the two species may have comparable functions or regulatory mechanisms. This provides significant insight into the conservation and divergence of the BRX gene family across plant species.
Figure 2.
Phylogenetic analysis of BRX gene family. Phylogenetic tree was constructed using MEGA11 with 1000 bootstrap replications based on a full-length amino acid sequence alignment of 40 high-fidelity PgERF proteins.
Figure 2.
Phylogenetic analysis of BRX gene family. Phylogenetic tree was constructed using MEGA11 with 1000 bootstrap replications based on a full-length amino acid sequence alignment of 40 high-fidelity PgERF proteins.
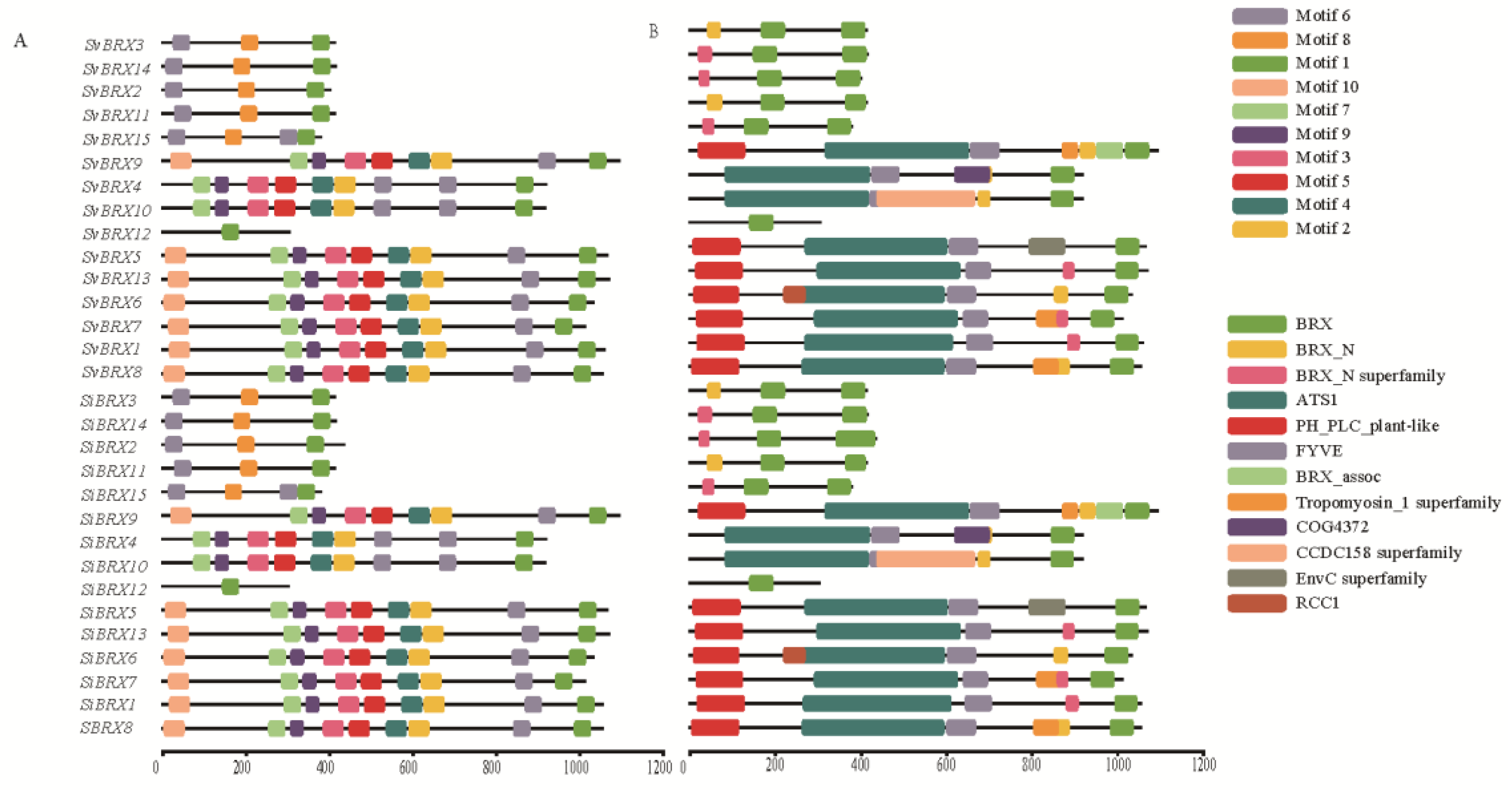
2.3. Conserved Motif and Domain Analyses of SiBRX Genes
This The motif analysis revealed the presence of 10 distinct motif types among the 30
BRX genes (
Figure 3A). Motif 1 was consistently present in all 30 genes. In contrast, Motif 6 was identified in 28
BRX genes, excluding
SiBRX12 and
SvBRX12. Additionally, 12 types of conserved domains were identified, 4 of which were specifically associated with the BRX domain: BRX, BRX_N, BRX_N superfamily, and BRX_assoc (
Figure 3B). The BRX domain was present in all 30
BRX genes, whereas the BRX_N and BRX_N superfamily domains appeared in only 10 genes. The BRX_assoc domain was exclusively found in
SiBRX9 and
SvBRX9. The diverse domain repertoire observed in the BRX gene family suggested functional diversity, evolutionary advantage, and a complex regulatory network. These findings provide a strong theoretical foundation for further investigations into the practical and evolutionary roles of the BRX gene family across different species.
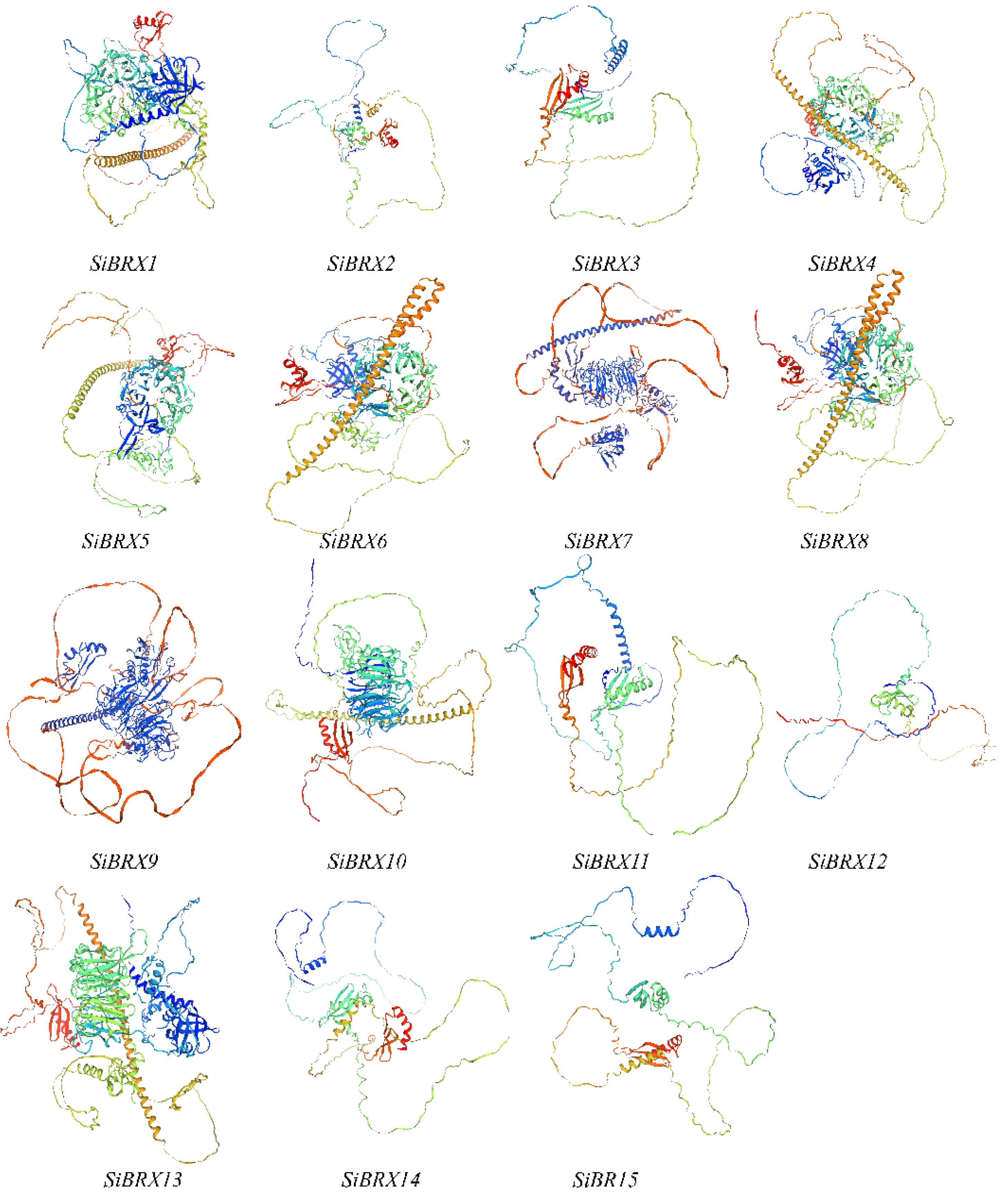
2.4. Estimating the 3D Structures of SiBRX Proteins
The function of a gene is closely linked to the structure of the protein it encodes. Homology modeling is used to predict the three-dimensional (3D) structure of a protein based on its amino acid sequence. The 3D structures of 15 SiBRX proteins were predicted using experimentally determined structures as templates. As shown in
Figure 4, the 3D models of all 15 genes included both β-sheets and α-helices. The 3D structures of SiBRX1, SiBRX4, SiBRX5, SiBRX6, SiBRX7, SiBRX8, SiBRX9, SiBRX10, and SiBRX13 included a ring-like structure composed of β-folds, and all SiBRX protein structures were surrounded by disordered loop structures.
2.5. miRNA Prediction
In this study, we focused on exploring the interaction between genes and miRNAs in foxtail millet. Through a series of rigorous bioinformatics predictions and experimental validations, we successfully identified four genes in foxtail millet that exhibit interactions with miRNAs from rice. The first interaction pair is SiBRX2 and osa - miR5801. Our analysis clearly demonstrated a significant interaction between them. Similarly, SiBRX5 was found to interact with osa - miR5075, SiBRX11 with osa - miR2927, and SiBRX14 with osa - miR396a - 3p.
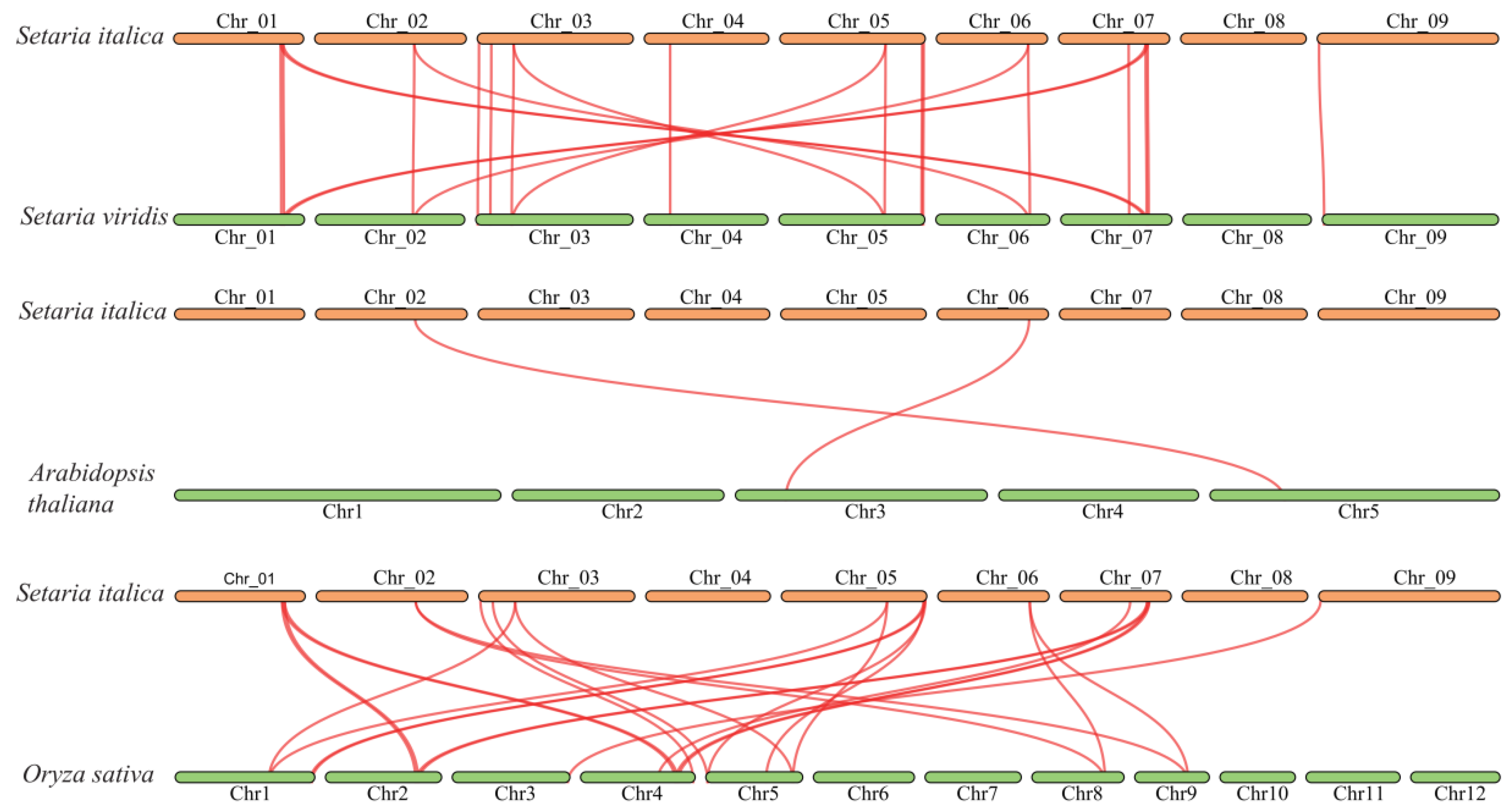
2.6. Gene Duplication and Collinearity Analysis of SiBRX Genes
This study investigated the evolutionary history of the foxtail millet SiBRX family and its homologous relationships with other species. The selection pressures acting on these genes during evolution were assessed by calculating the Ka/Ks ratios between corresponding
BRX genes in green foxtail and foxtail millet. The results showed that the Ka/Ks ratios for 15 pairs of duplicated genes were all less than 1, with some gene pairs having Ka or Ks values of zero (
Table S2). This finding indicated that
BRX genes were highly conserved during the evolution from green foxtail to foxtail millet, suggesting that these genes may have undergone purifying selection to maintain the stability of their essential roles. Additionally, the homologous
BRX genes among foxtail millet, green foxtail, Arabidopsis, and rice were examined. Further, 20, 2, and 22 pairs of homologous genes were identified between foxtail millet and green foxtail, foxtail millet and Arabidopsis, and foxtail millet and rice, respectively, through sequence alignment. Identifying these homologous genes established a basis for exploring the evolution and functional diversity of the BRX gene family among different plant species (
Figure 5).
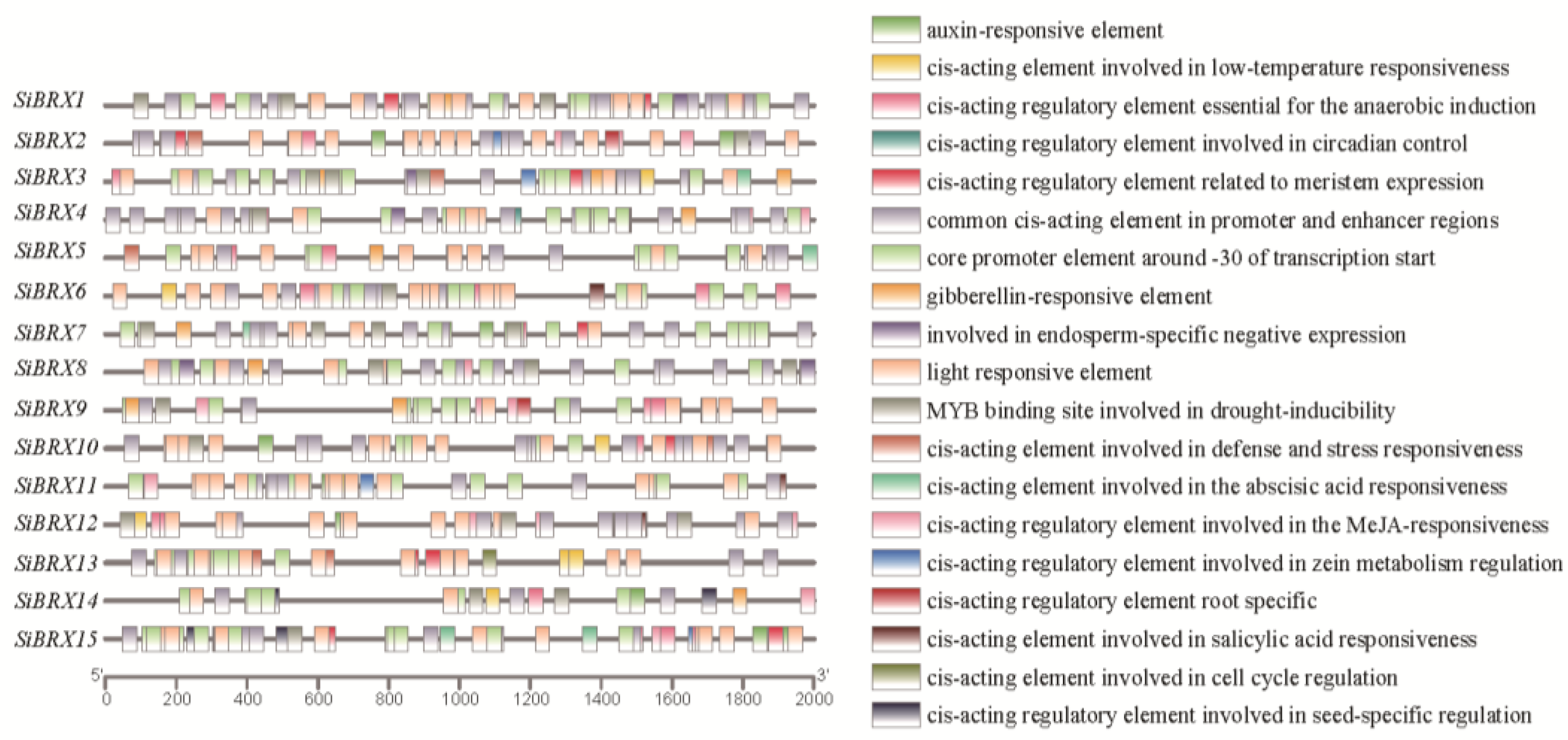
2.7. Analysis of Cis-Regulatory Elements in SiBRX Genes
To explore the roles and regulatory mechanisms of SiBRX genes, an in-depth examination was conducted on cis-acting elements within their promoter regions. Using PlantCARE software, these elements were identified and classified in the 2000-bp upstream region of the start codons for the
SiBRX genes (
Figure 6). The elements were grouped into four categories: stress-responsive, light-responsive, hormone-responsive, and growth and development-related elements. Notably, stress-responsive elements were prevalent, including low-temperature-responsive elements and drought-stress-responsive elements (MBSs), across the promoters. Hormone-responsive elements showed diversity, comprising gibberellin-responsive elements, abscisic acid–responsive elements (ABREs), jasmonic acid–responsive elements (TGACG motifs), salicylic acid–responsive elements (TCA elements), and auxin-responsive elements (TGTCTC motifs). These findings indicated that the
SiBRXs might regulate plant hormone signaling and abiotic stress responses. Significant variations were observed in the presence of hormone- and stress-responsive elements among
SiBRX genes. For instance, 13 of the 15
SiBRX genes contained auxin-responsive elements, whereas
SiBRX3,
SiBRX4,
SiBRX5,
SiBRX8,
SiBRX9, and
SiBRX13 lacked these elements.
SiBRX7 contained three auxin-responsive elements in its promoter region. This variation highlights the potential distinct roles of different
SiBRX genes in plant hormone and stress signaling pathways. Overall, the diverse
cis-acting elements in SiBRX promoters provide insights into their regulatory functions, supporting their involvement in plant physiological responses and laying a foundation for further exploration of their roles in the growth, development, and environmental adaptation of foxtail millet.
2.8. Expression Patterns of SiBRX Genes
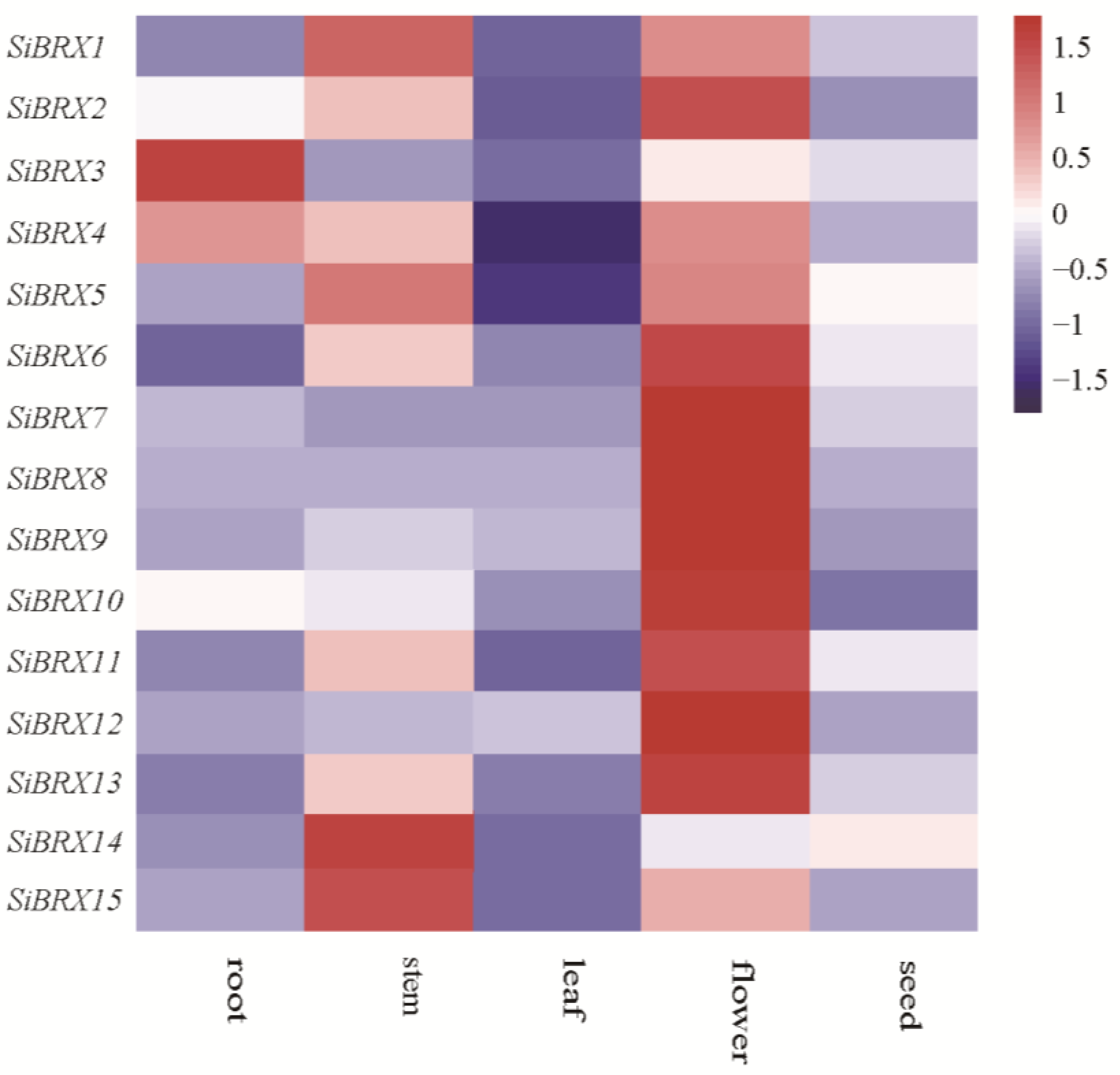
This study analyzed the expression of
SiBRXs in roots, stems, leaves, flowers, and seeds of foxtail millet using publicly available transcriptomic data and generated a heatmap (
Figure 7). The heatmap revealed distinct tissue-specific expression patterns among
SiBRX genes. Specifically,
SiBRX1,
SiBRX5,
SiBRX14, and
SiBRX15 were highly expressed in the stem, whereas
SiBRX2,
SiBRX6,
SiBRX7,
SiBRX8,
SiBRX9,
SiBRX10,
SiBRX11,
SiBRX12, and
SiBRX13 showed high expression in flowers.
SiBRX3 and
SiBRX4 exhibited elevated expression in roots. All
SiBRX genes demonstrated high expression levels in seeds. These results suggested crucial roles of
SiBRX genes in multiple tissues in millet, with their expression indicating high tissue specificity.
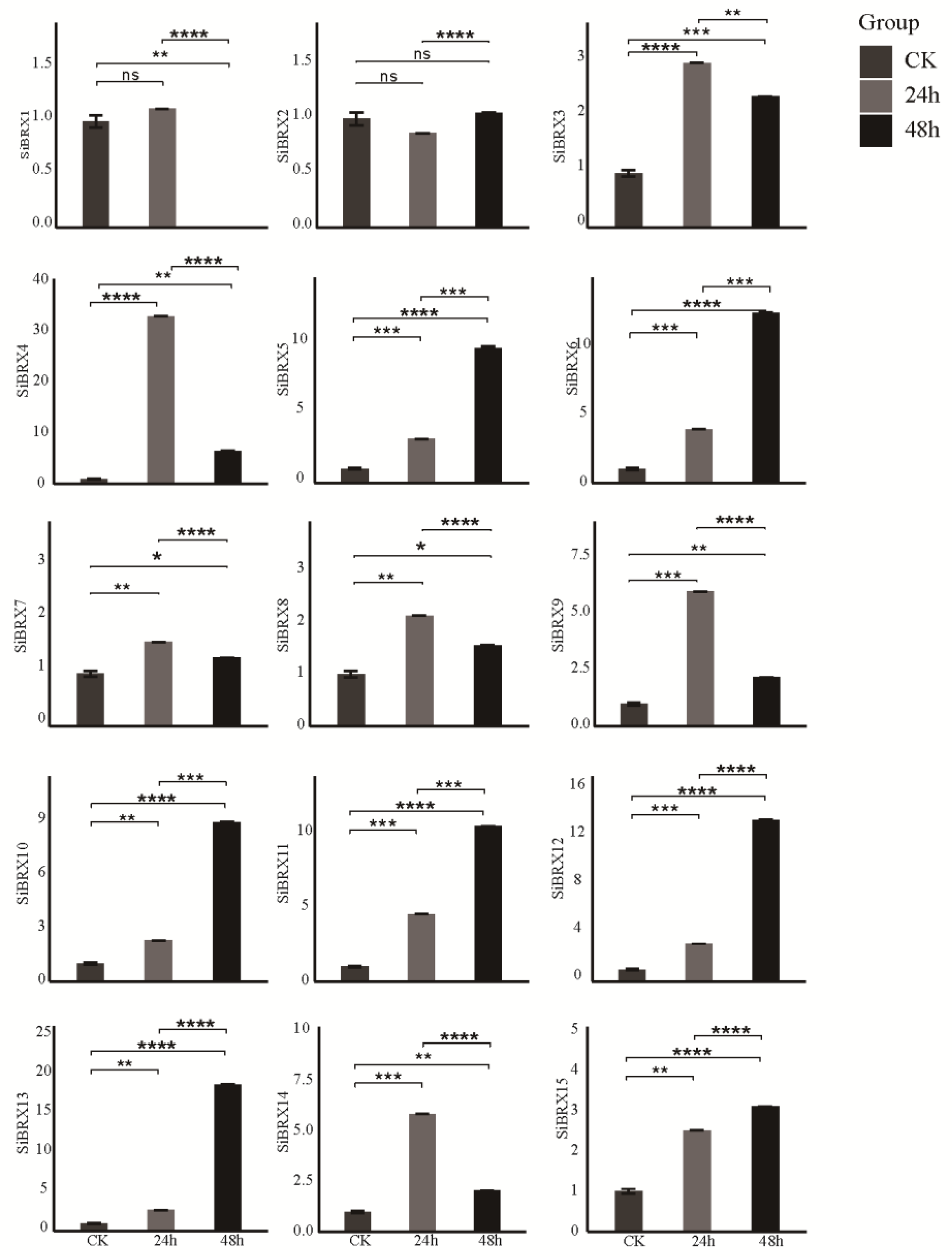
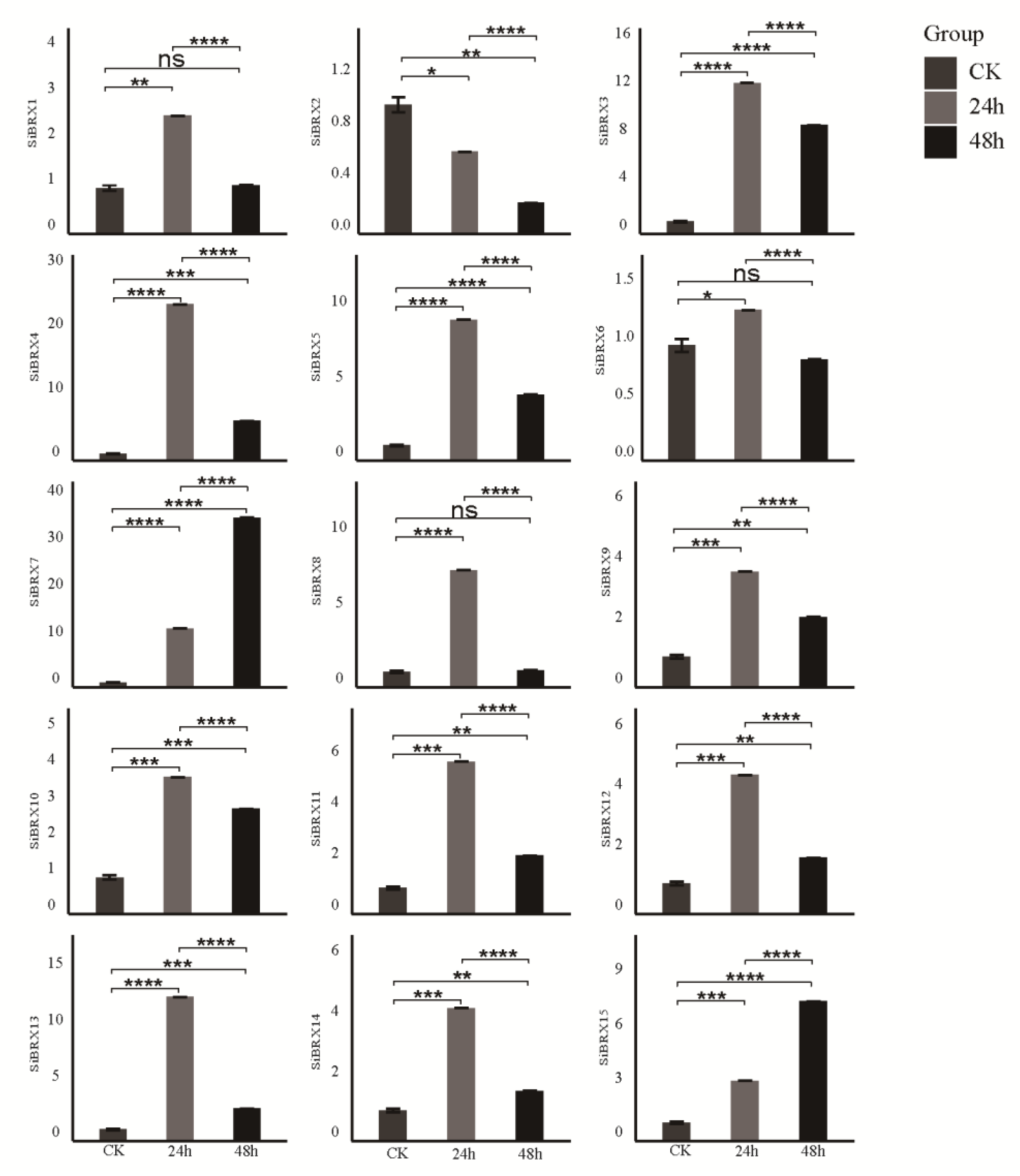
2.9. Analysis of SiBRX Expression Under Different Abiotic Stress Conditions
This study further explored the role of
SiBRX genes in stress resistance using 14-day-old seedlings of "Jingu 21”and subjecting them to different abiotic stress treatments, including cold stress, NaCl-induced salt stress, and PEG-induced drought stress. The transcriptional expression patterns of all
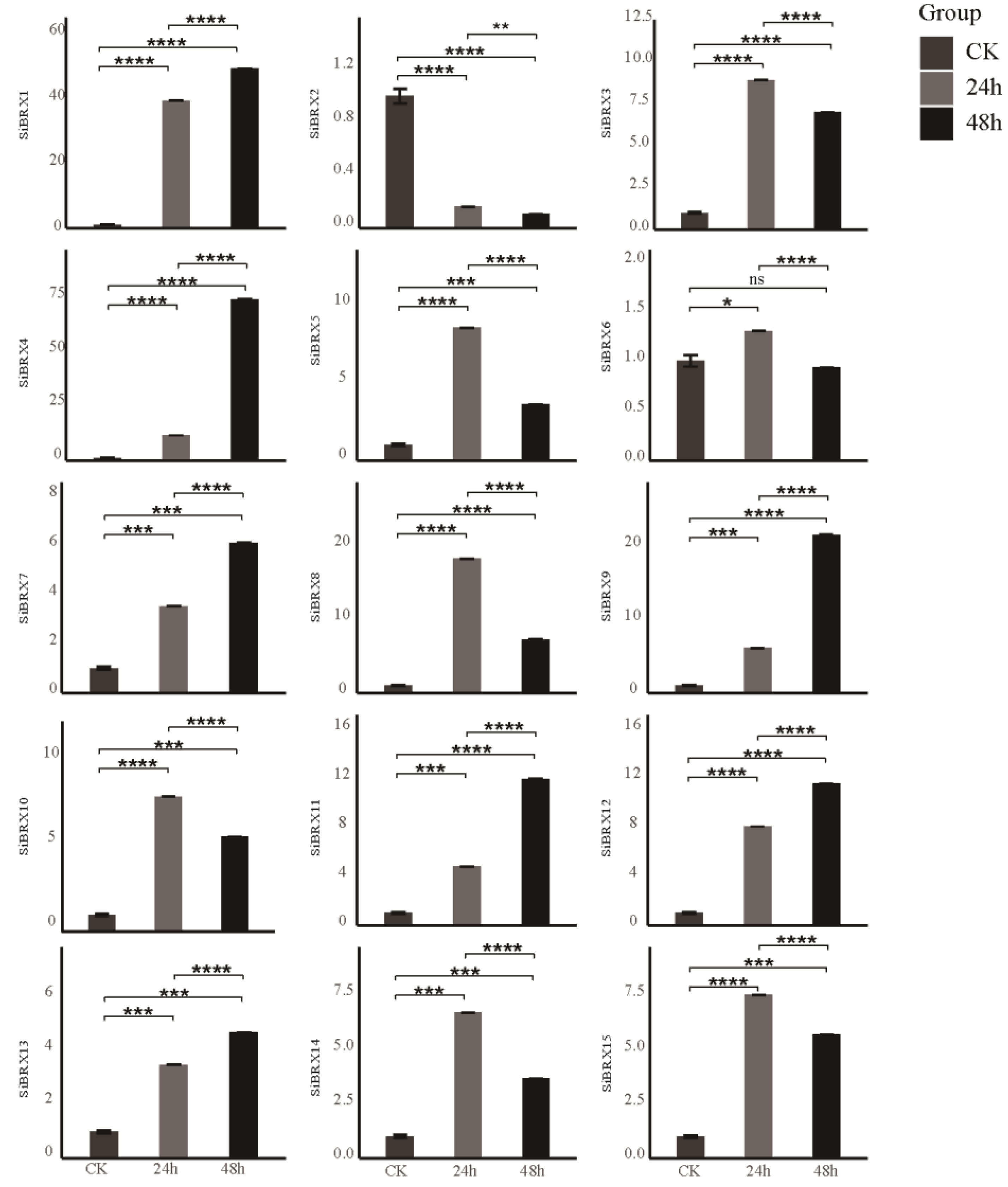
SiBRX genes were assessed using qRT-PCR to evaluate their response characteristics under different stress conditions. Under cold stress (
Figure 8), all genes except
SiBRX2 showed significant upregulation 24 h after treatment. In contrast,
SiBRX2 expression significantly decreased in both 24 and 48 h.
SiBRX1,
SiBRX4,
SiBRX7,
SiBRX9,
SiBRX11,
SiBRX12, and
SiBRX13 continued to show significant increases in expression in 48 h, compared with 24 h, whereas
SiBRX3,
SiBRX5,
SiBRX6,
SiBRX8,
SiBRX10,
SiBRX14, and
SiBRX15 exhibited significant decreases. Notably,
SiBRX6 expression fell below its pre-stress level. All genes except
SiBRX2 displayed significantly increased expression 24 h after drought stress treatment (
Figure 9).
SiBRX1,
SiBRX5,
SiBRX6,
SiBRX10,
SiBRX11,
SiBRX12,
SiBRX13, and
SiBRX15 showed further significant increases whereas
SiBRX3,
SiBRX4,
SiBRX7,
SiBRX8,
SiBRX9, and
SiBRX14 exhibited decreased expression in 48 h. However, despite these reductions, the expression levels remained significantly higher than those before stress treatment.
SiBRX2 did not exhibit significant changes in expression at any time point. All genes except
SiBRX2 were significantly upregulated 24 h after salt stress treatment (
Figure 10).
SiBRX7 and
SiBRX15 continued to increase significantly, whereas
SiBRX1,
SiBRX3,
SiBRX4,
SiBRX5,
SiBRX6,
SiBRX8,
SiBRX9,
SiBRX10,
SiBRX11,
SiBRX12,
SiBRX13, and
SiBRX14 showed significant decreases in 48 h. Specifically, the expression levels of
SiBRX1 and
SiBRX8 in 48 h were not significantly different from pre-stress levels, whereas the expression level of
SiBRX6 was significantly lower than that before stress. Additionally,
SiBRX2 expression continued to decline throughout the salt stress treatment.
3. Discussion
3.1. Identification and Analysis of SiBRX Genes in Foxtail Millet
This study embarked on a comprehensive exploration of the foxtail millet genome, leveraging state - of - the - art genomic sequencing and bioinformatics analysis techniques. Through meticulous data mining and in - depth computational analysis, we successfully identified 15 members belonging to the BRX gene family. This discovery is a significant step forward as it offers a more complete picture of the gene family’s presence in foxtail millet compared to previous, less comprehensive studies. Subsequently, a detailed phylogenetic analysis was carried out on these genes. Unlike traditional phylogenetic methods that rely on a limited set of markers, our analysis employed advanced algorithms and incorporated a broader range of genomic information. The findings were truly remarkable. We found that these genes could be categorized into various sub - families, suggesting an evolutionary functional diversification of the BRX genes. This discovery is not only consistent with previous studies on A. thaliana and rice but also adds a new layer of understanding. Our high - resolution phylogenetic tree revealed previously unrecognized evolutionary branches and divergence points, highlighting the unique evolutionary trajectory of the BRX gene family in foxtail millet. For instance, while it was known that AtBRX1 and AtBRX2 play crucial roles in root development, and AtBRX3 is involved in cell wall synthesis [
21,
22,
23], our analysis suggested that the corresponding genes in foxtail millet may have undergone further functional specialization during evolution, adapting to the specific ecological and physiological requirements of foxtail millet. Further analysis of protein domain features was conducted using the latest structural biology and bioinformatics tools. We found that foxtail millet BRX proteins typically contained multiple conserved domains, such as the BRX domain, FYVE domain, and RCC1 domain. These domains are highly conserved across BRX proteins in various plant species, underscoring their functional significance. However, our in - depth analysis also revealed some subtle variations in the structure and sequence of these domains in foxtail millet compared to other plants. These variations may be the key to understanding the unique functions of foxtail millet BRX proteins and could potentially be targeted for genetic engineering to enhance the plant’s traits. To gain a deeper understanding of the regulatory mechanisms of BRX genes in foxtail millet, we employed cutting - edge techniques for analyzing the promoter regions of these genes for cis - acting elements associated with specific expression patterns. Our approach was more comprehensive than previous studies, covering a wider range of potential regulatory elements. The results showed that the regulatory regions of BRX genes in foxtail millet contain a diverse array of cis - acting elements, including those related to stress responses, hormonal signaling, and photoperiod regulation. The promoters of multiple BRX genes were particularly enriched in ABRE and drought - responsive elements (DRE/CRT), indicating that these genes may be regulated by ABA signaling and drought stress. Similar findings have been reported in Arabidopsis and rice, where the promoters of AtBRX1 and AtBRX2 also exhibit a high abundance of ABRE and DRE/CRT elements [
21,
22,
23]. However, our study also identified some novel cis - acting elements that have not been previously reported in other plants, suggesting the existence of unique regulatory mechanisms in foxtail millet. In summary, this study comprehensively elucidated the composition, evolutionary relationships, and potential functions of the foxtail millet BRX gene family by phylogenetic analysis, gene structure characterization, protein domain analysis, and promoter region cis - acting element analysis. These findings not only enhance the understanding of the BRX gene family in foxtail millet but also provide valuable foundational data for further functional research and genetic improvement. Our study, with its novel techniques and in - depth analysis, sets a new standard for studying gene families in plants and paves the way for future breakthroughs in crop improvement. Previous studies on these four rice miRNAs have revealed their crucial roles in rice development and metabolism. Osa - miR5801 has been reported to be involved in the regulation of rice growth and development processes, potentially influencing key physiological pathways such as cell division and differentiation. Osa - miR5075 is known to participate in the metabolic regulation of rice, playing a role in maintaining the balance of various metabolites within the plant. Osa - miR2927 has been shown to be associated with important developmental stages in rice, affecting traits such as plant height and tillering. And osa - miR396a - 3p has been documented to regulate the expression of genes related to rice stress responses and growth regulation.Given the well - established functional importance of these rice miRNAs in rice, the identified interactions between these miRNAs and the corresponding SiBRX genes in foxtail millet strongly suggest the high functionality of SiBRX2, SiBRX5, SiBRX11, and SiBRX14 in foxtail millet. These findings not only provide new insights into the regulatory network of foxtail millet genes but also offer potential targets for future genetic improvement and breeding programs in foxtail millet.It is important to note that further in - depth studies are required to fully understand the specific molecular mechanisms underlying these interactions and how they contribute to the overall growth, development, and stress responses of foxtail millet. However, the results presented here lay a solid foundation for future research in this area.
3.2. Evolution of SiBRX Genes
This study comprehensively explored the evolutionary background of the SiBRX gene family in foxtail millet and its homologous associations with various other species. Building on the foundation of earlier genomic analyses of foxtail millet, which first unveiled the dynamics of gene family expansion and contraction within this particular species [
24], this research not only delved deeper into the BRX gene family but also incorporated cutting - edge bioinformatics approaches and emerging concepts in plant evolutionary genomics. In this investigation, we aimed to reveal the conserved features and the selective pressures the BRX gene family had endured during evolution from a multi - dimensional perspective. For instance, we employed advanced phylogenetic reconstruction algorithms that take into account complex evolutionary scenarios such as horizontal gene transfer and gene conversion events, which have been rarely considered in previous studies of the BRX gene family. Specifically, we examined the connection between green foxtail and foxtail millet. Based on previous findings of the high genomic similarity between these two species [
25], we further explored the micro - evolutionary changes at the nucleotide and amino - acid levels. By using single - nucleotide polymorphism (SNP) analysis and haplotype reconstruction, we were able to identify unique genetic signatures within the BRX genes that might contribute to the subtle phenotypic differences between green foxtail and foxtail millet. The analysis revealed high levels of conservation in BRX genes across these closely related species. The evolutionary rates of these genes were assessed by calculating the Ka/Ks ratios. To add more depth to this analysis, we also performed a sliding - window analysis of the Ka/Ks ratios along the gene sequences. This allowed us to detect regions within the BRX genes that might have experienced different selective pressures. The results indicated that the Ka/Ks ratios for 15 pairs of genes were <1, with some pairs even exhibiting zero values for either Ka or Ks. This strongly suggested that BRX genes underwent strong purifying selection during the evolutionary transition from green foxtail to foxtail millet, ensuring their critical functions in plant growth and stress responses. Furthermore, this study compared and identified homologous genes between BRX genes of foxtail millet and those of A. thaliana and rice. In addition to the traditional sequence - based homology search, we integrated structural genomics data. By comparing the three - dimensional protein structures of BRX genes from different species, we were able to identify conserved structural motifs that are likely to be functionally important. Through this comprehensive comparative analysis, the study identified 20, 2, and 22 pairs of homologous genes corresponding to the relationships between foxtail millet and green foxtail, foxtail millet and A. thaliana, and foxtail millet and rice, respectively. This cross - species identification not only enhances the comprehension of the evolutionary path of the BRX gene family but also highlights the functional conservation and common evolutionary traits of BRX genes within the Poaceae family. The high number of homologous genes between foxtail millet and green foxtail further confirms the close evolutionary ties between these two species. In contrast, the substantial number of homologous genes between foxtail millet and rice reaffirms the high conservation of BRX genes within the Poaceae family. Moreover, our discovery of specific structural motifs shared among these homologous genes provides new insights into the functional conservation mechanism at the molecular level, which has not been reported in previous studies of the BRX gene family. Overall, this study presents a more comprehensive and in - depth exploration of the SiBRX gene family, integrating multiple omics data and advanced analytical methods, which significantly enriches our understanding of the evolution and function of this important gene family
3.3. Spatio-Temporal Patterns of Expression for SiBRX Genes and Their Responses to Abiotic Stress
This study thoroughly explored the specificity of expression within the SiBRX gene family across various tissues of foxtail millet and the mechanisms of their responses to different abiotic stress factors. In the realm of current plant genetics research, while the general understanding of gene families related to plant stress - responses has been advancing, the in - depth exploration of the SiBRX gene family in foxtail millet, a vital cereal crop with unique physiological characteristics and ecological adaptability, still holds a wealth of uncharted territory. The study indicated that different SiBRX genes exhibited significant expression differences in different tissues of foxtail millet, which were closely related to their specific mechanisms during plant growth. These tissue - specific expression patterns are not only a basic manifestation of gene regulation but also imply a high - level coordination of the SiBRX gene family in different physiological processes of foxtail millet. For example, in the meristematic tissues, certain SiBRX genes may be highly expressed to promote cell division and differentiation, which is crucial for the establishment of the plant’s body structure. In contrast, in the mature photosynthetic tissues, other SiBRX genes may play roles in optimizing photosynthetic efficiency and carbon metabolism. Such tissue - specific fine - tuning of gene expression represents a sophisticated genetic regulatory network that has not been fully explored in previous studies on related gene families. The results provide important perspectives on the functional variety within the BRX gene family. To date, the BRX gene family has been a subject of research in some plant species, but the diversity of functions within the family, especially in foxtail millet, remains to be fully uncovered. The SiBRX gene family in foxtail millet may have evolved unique functions due to its long - term adaptation to specific ecological environments. While many studies have focused on well - known stress - response genes, the SiBRX gene family in foxtail millet may offer new insights into stress - resistance mechanisms. The findings revealed that most SiBRX genes exhibited a significant upregulation after 24 h of exposure to cold stress, suggesting their essential role in the rapid adaptive response to low - temperature conditions. This rapid upregulation may be related to the activation of a series of cold - responsive signal transduction pathways. For instance, SiBRX genes may interact with cold - induced transcription factors to promote the expression of downstream cold - tolerance genes, which is a novel regulatory mechanism that has not been well - documented in other gene families. SiBRX2 expression was significantly downregulated under cold stress, possibly related to its unique regulatory mechanisms or involvement in specific physiological processes. This downregulation could be a strategic adjustment by the plant to re - allocate resources during cold stress. For example, SiBRX2 may be involved in processes that are energy - consuming under normal conditions, and its downregulation during cold stress helps the plant conserve energy for more essential cold - resistance activities. The expression of most SiBRX genes, except SiBRX2, was generally upregulated within 24 h under drought stress, indicating their participation in regulating drought resistance. This differential expression pattern among SiBRX genes suggests a division of labor within the gene family during drought stress. Some genes may be responsible for maintaining cell turgor, while others may be involved in antioxidant defense systems. This complex and coordinated response mechanism is a new discovery in the study of plant drought - resistance genes. The expression levels started declining after 48 h under salt stress, indicating the complex adaptive strategies and dynamic regulatory mechanisms of plants under prolonged salt stress. This decline in gene expression may be a result of the plant’s attempt to avoid over - activation of stress - response pathways, which could lead to excessive energy consumption and potential damage to the plant. The SiBRX gene family may be involved in a feedback regulatory loop that precisely controls the intensity and duration of the stress - response process. SiBRX2 exhibited a downregulation trend under all three conditions, indicating that this gene may play unique regulatory roles in stress responses. Considering its tissue - specific expression patterns, the low expression of SiBRX2 may be associated with its protective or inhibitory roles in certain stress conditions or specific tissues. For example, in the root tissue under salt stress, the downregulation of SiBRX2 may prevent excessive ion uptake, thus protecting the plant from salt - induced toxicity. The investigation of SiBRX gene family expression across various tissues and their varying responses to a range of stress conditions in this study uncovered potential mechanisms influencing the growth, development, and stress responses of foxtail millet. The expression pattern of SiBRX2 under stress conditions provides new directions for research, suggesting that its function in specific stress or tissue conditions may be distinct. These findings not only improve the understanding of the role of the BRX gene family in plant adaptive evolution but also provide essential target genes for genetic improvement in foxtail millet and other grass crops, particularly for developing new varieties with enhanced stress tolerance. Further studies are needed to investigate the functions and mechanisms of individual members of the SiBRX family so as to provide more precise molecular tools for improvement in crop stress resistance. Future research could focus on the interaction between SiBRX genes and other key stress - response genes, as well as the post - translational modification of SiBRX proteins, which may further uncover the hidden complexity of the SiBRX gene family in foxtail millet’s stress - response system.
4. Materials and Methods
4.1. Identification and Analysis of BRX Family Genes in Millet
The analysis utilizing BLASTP was performed with acid sequences derived from Arabidopsis and rice to pinpoint BRX family genes within foxtail millet, applying an E-value cutoff of 10⁻⁵ [
26]. This investigation was executed utilizing genomic references of green foxtail (Setaria viridis v2.1) and foxtail millet (Setaria italica v2.2) available in the Phytozome database (
https://phytozome-next.jgi.doe.gov; accessed December 1, 2024) [
27]. Moreover, the existence of the BRX domain (PF08381; E-value cutoff of 0.001) [
28] within these sequences was further validated using PFAM 37.0 (
https://pfam.xfam.org; accessed on December 1, 2024) [
29]. Furthermore, various physical and chemical characteristics of the foxtail millet BRX proteins, such as amino acid composition, theoretical isoelectric point, molecular weight, aliphatic index, instability index, and grand average of hydropathicity, were calculated using ExPASy (
https://www.expasy.org; accessed December 1, 2024). The prediction of subcellular localization for
SiBRXs and
SvBRXs was performed through the WoLF PSORT tool (
https://www.genscript.com/wolf-psort.html; accessed on December 1, 2024) [
30]. The members of the foxtail millet BRX gene family were designated according to their chromosomal arrangement.
4.2. Analysis of Phylogeny and Chromosomal Positioning of BRX Family Genes
The BRX protein sequences from rice, Arabidopsis, green foxtail, and foxtail millet were aligned using ClustalW (2.0) to illuminate the evolutionary relationships amid BRX-like genes [
31]. A phylogenetic tree was then constructed using the neighbor-joining (NJ) technique with the assistance of MEGA 11 software [
32], setting the bootstrap confidence level at 1000 iterations. This tree was visualized through the iTOL online platform (
http://itol.embl.de; accessed on December 1, 2024) [
33]. Additionally, genomic annotation files and the Basic Circos program in TBtools v2.003 [
34] were used to map and visualize the identified BRX gene family members on the chromosomes of the foxtail millet genome.
4.3. Analysis of Conserved Domains and Motifs
The analysis of conserved domains for the SiBRX gene family was conducted via the NCBI Batch CD-Search (
https://www.ncbi.nlm.nih.gov; accessed on December 1, 2024) [
35] and visualized with TBtools software. For motif analysis, the SiBRX protein amino acid sequences were submitted to the MEME online platform (
https://meme-suite.org/meme; accessed on December 1, 2024) [
36], with a specification of a maximum of 10 motifs. The motifs were sequentially named from Motif 1 to Motif 10 based on their arrangement from the N-terminus to the C-terminus.
4.4. Cis-Acting Regulatory Elements and miRNA Prediction
Promoter sequences for the BRX gene family, situated 2 kb upstream of the ATG start codon in foxtail millet, were obtained from the Phytozome database. An analysis of
cis-acting elements was performed with PlantCARE software [
37], and the results were ultimately presented with TBtools software. cDNA sequences of the TaEs were used as candidate targets for predicting potential miRNAs. Using the psRNATarget Server (
https://www.zhaolab.org/psRNATarget/) with default parameters, we then searched for candidate targets by screening the mature sequences of rice miRNAs obtained from the miRbase database (
http://www.mirbase.org/)
4.5. Synteny Analysis and Calculation of Nonsynonymous to Synonymous Substitution Ratios
Both intragenomic and intergenic synteny analyses were conducted for the BRX gene family in Arabidopsis, foxtail millet, rice, and green foxtail using MCScanX 3.0 [
38]. The results were visualized with TBtools software. Homologous gene pairs were identified depending on their location and sequence similarities. The nonsynonymous to synonymous substitution (Ka/Ks) ratios were calculated using the codeml program from the PAML package to assess molecular selective pressures [
39]. Genes with Ka and Ks values of zero were excluded from the analysis to avoid errors due to sequence saturation or misalignment. Genes with Ka/Ks ratios >1 were classified as positively selected genes, whereas those with ratios <1 were considered negatively selected genes.
4.6. Forecasting the Three-Dimensional Structure of the SiBRX Protein
The SiBRX protein structures were modeled through homology using SWISS-MODEL (
https://swissmodel.expasy.org; accessed on December 1, 2024) [
4][
44]. These models were developed based on template structures that exhibited over 30% sequence identity. The quality assessment of the predicted protein models was conducted with SAVES v6.0 (
https://saves.mbi.ucla.edu; accessed on December 1, 2024) [
45], ensuring that only templates meeting at least three quality criteria were included in the final model. Visualization of the protein structures was accomplished via VMD (
http://www.ks.uiuc.edu; accessed on December 1, 2024) [
46].
4.7. Expression Profiling of SiBRX Genes
The expression data for various patterns of the
SiBRXs were obtained from the EMBL-EBI database (
https://www.ebi.ac.uk; dataset GEO-DS36391; accessed on December 1, 2024). Tissue-specific expression profiling and heatmap visualization were conducted using the "pheatmap" package in R Studio.
4.8. Plant Resources and Treatment Approaches
Jingu 21 foxtail millet seeds were germinated in a room at 26°C with 65% humidity under an 8-h dark/16-h light cycle. After 2 days, the seedlings were moved to plastic containers filled with a diluted Hoagland's nutrient solution at half strength to promote additional growth [
35]. For cold and salt stress treatments, 12-day-old seedlings were subjected to either cold (4°C) or 150µM NaCl treatment. For drought stress, a 20% PEG solution was applied at the seedling stage. Leaf samples were collected at 24 and 48 h post-treatment, with seedlings grown at 26°C serving as the control group. All samples were immediately flash-frozen in liquid nitrogen and stored at –80°C for further analysis.
4.9. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction
Leaf tissues were processed to extract total RNA using the TRIzol reagent (Accurate Biology, Changsha, China). Following this, cDNA synthesis was achieved through the use of a reverse transcription kit from the same company (Accurate Biology). For conducting quantitative real-time polymerase chain reaction (qRT-PCR), the SYBR Green assay kit (Accurate Biology) was employed. The design of primers was carried out using Primer Premier 5 software, with relevant details available in
Table S1. The qRT-PCR reactions occurred on a Bio-Rad CFX system, utilizing a reaction mix of 20 µL that included 10 µL of 2× SYBR Green Pro Taq HS premix, 2 µL of cDNA, 0.4 µL of each forward and reverse primer, along with 7.2 µL of RNase-free water. The thermal cycling process began with an initial denaturation at 95°C for 30 s, which was followed by 40 cycles at 95°C for 5 s and 60°C for 30 s. SiActin (SETIT_026509mg) provided the internal control, and the relative expression levels of the targeted genes were calculated employing the 2−∆∆Ct method. A heatmap depicting the expression patterns of the
SiBRX gene was produced using TBtools, with expression values normalized through Log₂ transformation. To ensure reliability, each experiment was independently repeated a minimum of three times.
4.10. Statistical Analyses
To ensure reliability, each experiment was carried out in triplicate. The analysis of the data was conducted using SPSS 22, and the mean ± standard deviation for every experiment was computed. The Student t test was used to assess statistical significance, with P < 0.05 denoting statistical significance and P < 0.01 indicating highly significant differences.
5. Conclusions
This study identified 15 members of the BRX gene family within the millet genome and categorized them into 3 subfamilies through phylogenetic analysis. Members from the same subfamily displayed comparable conserved motifs and structural gene domains. An in-depth analysis of chromosomal distribution and gene duplication events indicated an uneven distribution of BRX genes throughout the chromosomes, highlighting evolutionary processes. The Ka/Ks analysis comparing BRX genes between green foxtail and foxtail millet affirmed the conservation of this gene family throughout the evolutionary shift from foxtail millet to green foxtail. Furthermore, the co-linearity analysis of BRX genes among green foxtail, foxtail millet, Arabidopsis, and rice illustrated the significant conservation of BRX genes within the grass family. Moreover, the identification of several stress-responsive and developmental regulatory elements within the BRX gene family implies their role in various biological processes, such as millet growth, development, and responses to abiotic stress. RT-qPCR assessments demonstrated that most members of the BRX gene family showed differential expression at the transcriptional level when exposed to abiotic stress conditions, though their exact functions warrant further exploration. Collectively, these findings offer a theoretical basis for upcoming investigations into the biological functions of the BRX gene family in millet and its evolutionary relatives, alongside their regulatory mechanisms regarding responses to abiotic stress.
Data Availability Statement The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org, Table S1. Target gene yeast double heterogeneous primer. Table S2. Ka/Ks values of homologous genes in foxtail millet and its ancestors.
Author Contributions
Conceptualization, XR, Y. methodology, XR, Y. software, XH, B.; validation, XH, B. formal analysis, XH, B. investigation, XR, Y. resources, J,Y. data curation, J,Y. writing—original draft preparation, XR, Y. writing—review and editing, XR, Y. visualization XR, Y.; and XH. B. supervision, ZJ,J. project administration, CY,W. funding acquisition, CY,W. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding
Acknowledgments
In this section, you can acknowledge any support given which is not covered by the author contribution or funding sections. This may include administrative and technical support, or donations in kind (e.g., materials used for experiments).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Xukai, Li.; Jianhua, Gao.; Jingyi, Song.; Kai, Guo.; Siyu, Hou.; Xingchun, Wang.; Qiang, He.; Yanyan, Zhang.; Yakun, Zhang.; Yulu, Yang.; Jiaoyan, Tang.; Hailang, Wang.; Staffan, Persson.; Mingquan, Huang.; Lishuai, Xu.; Linlin, Zhong.; Dongqin, Li.; Yongming, Liu.; Hua, Wu.; Xianmin, Diao.; Peng, Chen.; Xiaowen, Wang.; Yuanhuai, Han. Multi-omics analyses of 398 foxtail millet accessions reveal genomic regions associated with domestication, metabolite traits, and anti-inflammatory effects. Mol. Plant. 2022, 15(8): 1367-1383.
- Saxena, R.; Vanga, S.; Wang, J.; Orsat, V.; Raghavan, V. Millets for Food Security in the Context of Climate Change: A Review. Sustainability. 2018, 10, 2228. Author 1, A.; Author 2, B. Book Title, 3rd ed.; Publisher: Publisher Location, Country, 2008; pp. 154–196. [CrossRef]
- Chanwala, J.; Khadanga, B.; Jha, D.K.; Sandeep, I.S.; Dey, N. MYB Transcription Factor Family in Pearl Millet: Genome-Wide Identification, Evolutionary Progression and Expression Analysis under Abiotic Stress and Phytohormone Treatments. Plants. 2023, 12, 355.Author 1, A.B. (University, City, State, Country); Author 2, C. (Institute, City, State, Country). Personal communication, 2012. [CrossRef]
- Allah, W.; Jamshaid, H.; Tauqeer, A.Y.; Muhammad, A.; Muhammad, A.R. Foliar Application of Silicon Improved Physiological Indicators, Yield Attributes, and Yield of Pearl Millet (Pennisetum glaucum L.) Under Terminal Drought Stress. Journal of Soil Science and Plant Nutr. 2022, 22(4): 4458-4472.
- Zhang, J.; Wu, J.; Guo, M.; Aslam, M.; Wang, Q.; Ma, H.; Li, S.; Zhang, X.; Cao, S. Genome - wide characterization and expression profiling of Eucalyptus grandis HD-Zip gene family in response to salt and temperature stress. BMC Plant Biol. 2020, 20:451. [CrossRef]
- Lai, DL.; Yao, X.; Yan, J.; Gao, A.; Yang, H.; Xiang, DB.; Ruan, JJ.; Fan, Y.; Cheng, JP. Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in foxtail millet (Setaria italica). BMC Genomics. 2022, 23:549. [CrossRef]
- Zhang, X.; Zhou, Y.; Ding, L.; Wu, Z.; Liu, R.; Meyerowitz, EM. Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell. 2013, 25:83–101. [CrossRef]
- Santos, LA.; de-Souza, SR.; Fernandes, MS. OsDof25 expression alters carbon and nitrogen metabolism in Arabidopsis under high N-supply. Plant Biotechnol Rep. 2012, 6:327–37.
- Gendron, JM.; Pruneda-Paz, JL.; Doherty, CJ.; Gross, AM.; Kang, SE.; Kay, SA. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA. 2012, 109: 3167–72.
- Du, J.; Miura, E.; Robischon, M.; Martinez, C.; Groover, A. The Populus Class III HD ZIP transcription factor POPCORONA affects cell differentiation during secondary growth of woody stems. PLoS ONE. 2011, 6: e17458. [CrossRef]
- Yang, B.; Jiang, Y.; Rahman, MH.; Deyholos, MK.; Kav, NN. Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol. 2009, 9: 68. [CrossRef]
- Ramírez, V.; Coego, A.; López, A.; Agorio, A.; Flors, V.; Vera, P. Drought tolerance in Arabidopsis is controlled by the OCP3 disease resistance regulator. Plant J. 2009, 60: 929. [CrossRef]
- Samuel, W.H.K,; Petra, M.; Surbhi, R.; Alina, G.; Bernard, M.E.M.; Alkistis, E.L. B.; Melina, Z.; Martina, K.; Ulrich, Z.H.; Claus, S.; Christian, S.H. Mapping and engineering of auxin-induced plasma membrane dissociation in BRX family proteins. Plant Cell. 2021, 33 (6): 1945-1960.
- Georgette, C.; Briggs, C.F.; Mouchel, S.H. Characterization of the Plant-Specific BREVIS RADIX Gene Family Reveals Limited Genetic Redundancy Despite High Sequence Conservation. Plant Physiol. 2006, 140(4): 1306-1316.
- Céline, F.M.; Karen, S.O.; Christian, S.H. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature. 2006, 443(7110): 458-461.
- Sneha, T.; Senthilkumar, M.; Priyanka, R.; Monika, D. Genome wide analysis of BREVIS RADIX gene family from wheat (Triticum aestivum): A conserved gene family differentially regulated by hormones and abiotic stresses. Int. J. Biol. Macromol. 2023, 232(0): 123081-123081.
- Linsan, L.; Sarah, J.; Chiara, C.; Micha, B.; Miguel, A.S.; Trisha, M.; Yue, Z.; Mhmoud, E.; Linda, M.; Miriam, S.; Thomas, E.B.; Ian, D.B.; Luke, R.; Penny, W.K.; Robbie, W.; Alistair, M.H.; Sarah, M.M. Conserved signalling components coordinate epidermal patterning and cuticle deposition in barley[J].Nat. Commun. 2022, 13.
- Yoshiaki, I.; Masaichi, M.; Yasuo, N.; Hidemi, K.; Akira, Y. Characterization of Rice Mutants Deficient in the Formation of Crown Roots. Breed. Sci. 2001, 51(2): 123-129.
- Gábor, K.; Balázs, T.; T, B.; Gabriella, S.; Andrea, J.; Gábor, G. Glutathione reductase activity and chilling tolerance are induced by a hydroxylamine derivative BRX-156 in maize and soybean. Plant Sci. 2001, 160(5): 943-950.
- Zhang, YY.; Liang, JS.; Cai, XH.; Chen, HX.; Wu, J.; Lin RM.; Cheng, F.; Wang, XY. Divergence of three BRX homoeologs in Brassica rapa and its effect on leaf morphology. Hortic. Res. 2021, 8(1): 0-0. [CrossRef]
- Chikako, S.; Giorgina, B.; Christian, S. Intraspecific competition reveals conditional fitness effects of single gene polymorphism at theArabidopsisroot growth regulatorBRX. New Phytol. 2008, 2, 180(1): 71-80.
- Julien, B.; Emanuele, S.; Danuše, T.; Laura, Ragni.; Miroslav, S.; Christian, S.H. BRX promotes Arabidopsis shoot growth. New Phyto. 2010, 2, 188(1): 23-29.
- Liu, J.; Liang, DH.; Song, Y.; Xiong, LZ. Systematic identification and expression analysis of BREVIS RADIX-like homologous genes in rice. Plant Sci. 2010, 1, 178(2): 183-191. [CrossRef]
- He, QW.; Wang, CC.; He, QW.; Zhang, J.; Liang, HK.; Lu, ZF.; Xie, K.; Tang, D.; Zhou, YZ.; Liu, B.; Zhi, H.; Jia, GQ.; Guo, GG.; Du, HL.; Diao XM. A complete reference genome assembly for foxtail millet and Setaria-db, a comprehensive database for Setaria. Mol. Plant. 2023, 1, 0(0): 0-0. [CrossRef]
- He, Q.; Tang, S.; Zhi, H.; Chen, JF.; Zhang, F.; Liang, HK.; Ornob, A.; Li, HB.; Zhang, H.; Xing, LH.; Li, xkWei Zhang,Hailong Wang,Junpeng Shi,Huilong Du,Hongpo Wu,Liwei Wang,Ping Yang,Xing Lü,Hongshan Yan,Zhongqiang Song,Jinrong Liu,Haigang Wang,Tian Xia,Qiao Zhang,Guojun Feng,Ruifeng Guo,Wei Zhu,Yi Ren,Hao Hu,Mingzhe Li,Aiying Zhang,Erhu Guo,Yilong Feng,Qingquan Li,Yanli Liu,Bohong Tian,Xing-Ming Zhao,Ruiling Jia,Baili Feng,Jiewei Zhang,Jianhua Wei,Jinsheng Lai,Guanqing Jia,Michael D. Purugganan,Xianmin Diao. A graph-based genome and pan-genome variation of the model plant Setaria Nature Genetics. 2023 Jun 8;55 (7):1232-1242. [CrossRef]
- Liŭ, D.; Cui, YJ.; Zhao, ZL.; Li, SY.; Liang, D.; Wang, CL.; Feng, G.; Wang, JH.; Liu, ZL. Genome-wide identification and characterization of the BES/BZR gene family in wheat and foxtail millet. BMC Genomics. 2021, 22(1): 0-0. [CrossRef]
- David, G.; Shu, SQ.; Russell, W.H.; Rochak, N.; Richard D.H.; Joni, F.; Therese, M.; William, D.; Uffe, H.; Nicholas, H.P.; Daniel, S.R. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–118.
- Jaina, M.; Sara, C.; Lowri, W.; Matloob, Q.; Gustavo, S.; Erik, L.L.; Silvio, C.E.; Lisanna, P.; Shriya, R.; Lorna, R.; Alex, B.; Alex, B. Pfam: The protein families database in 2021. Nucleic Acids Research. 2020. 49: D412-D419.
- Céline, F.; Mouchel, G.C.; Briggs, C.S.; Hardtke. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes & Development. 2004;18 (6):700-714.
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinform. 2006, 64, 643–651.
- Larkin, M. Clustal W and Clustal X v. 2.0. Bioinformatics. 2007, 23, 2947–2948.
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [CrossRef]
- Wang, JY.; Farideh, C.; Myra, K.D.; Noreen, R.G.; Marc, G.; Lu, G.; Gabriele, H.M.; Song, JY.; Narmada, T.; Roxanne, A.Y.; Yang, MJ.; Chen, Z.; Christopher, J.L.; Aron, MB. The conserved domain database in 2023 Nucleic Acids Research. 2022, 51(D1): D384-D388.
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [CrossRef]
- Rombauts, S.; Dehais, P.; Van Montagu, M.; Rouze, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Carlos Sanchez-DelBarrio, J.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [CrossRef]
- Lin, M.; Dong, Z.; Zhou, H.; Wu, G.; Xu, L.; Ying, S.; Chen, M. Genome-Wide Identification and Transcriptional Analysis of the MYB Gene Family in Pearl Millet (Pennisetum glaucum). Int. J. Mol. Sci. 2023, 24, 2484. [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [CrossRef]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [CrossRef]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and SwissPdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo-distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771.
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017, 7, 10480. [CrossRef]
- Pontius, J.; Richelle, J.; Wodak, S.J. Deviations from Standard Atomic Volumes as a Quality Measure for Protein Crystal Structures. J. Mol. Biol. 1996, 264, 121–136. [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).