Submitted:
19 March 2025
Posted:
19 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Genomic DNA Extraction
2.2. Library Preparation and Sequencing
2.3. Pre-Processing, Read Filtering, Mitogenome Assembly, and Annotation
2.4. Nucleotide Sequence Composition Analysis
2.5. Phylogenetic Analysis
3. Results and Discussion
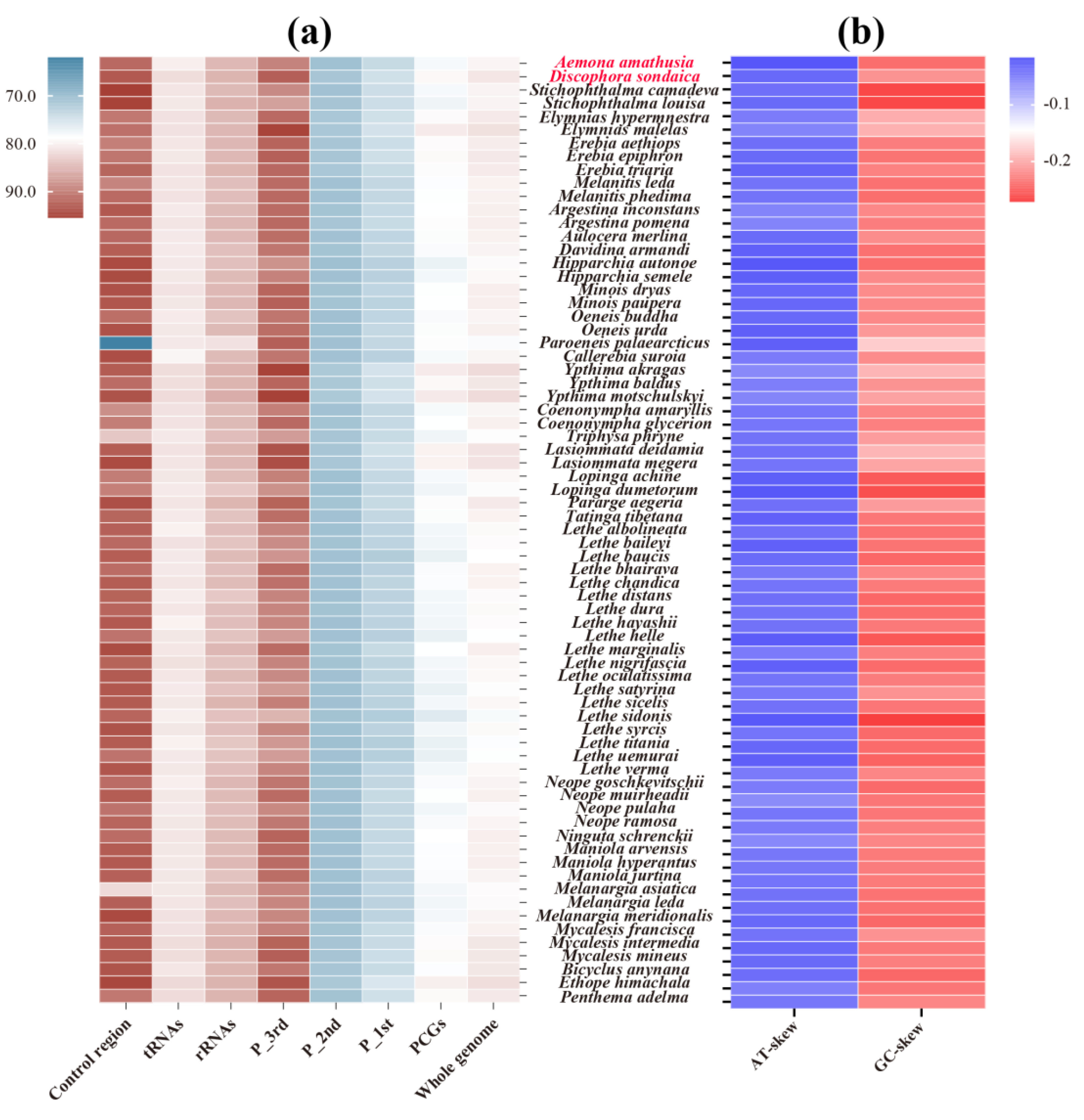
3.1. Mitogenome Organization and Base Composition
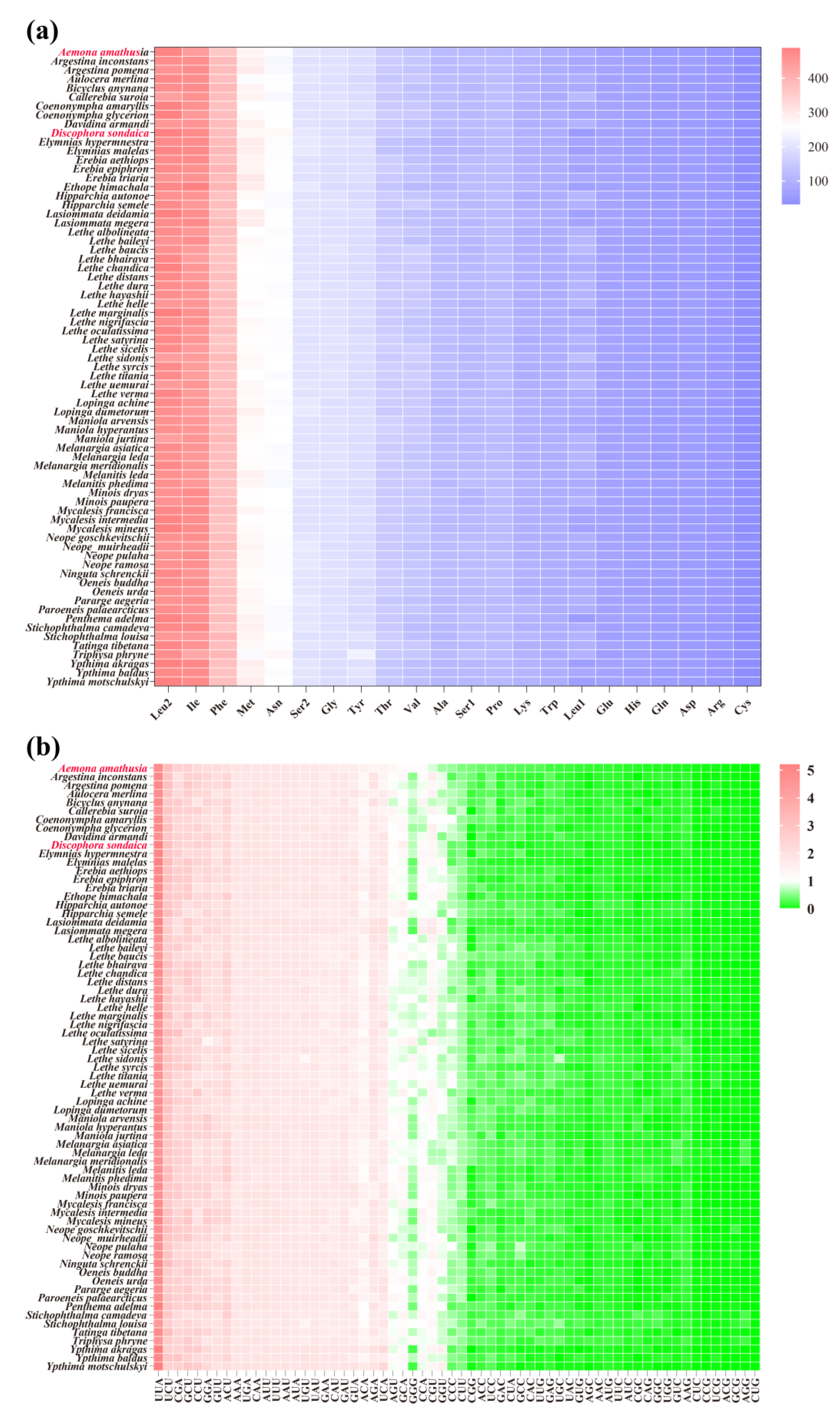
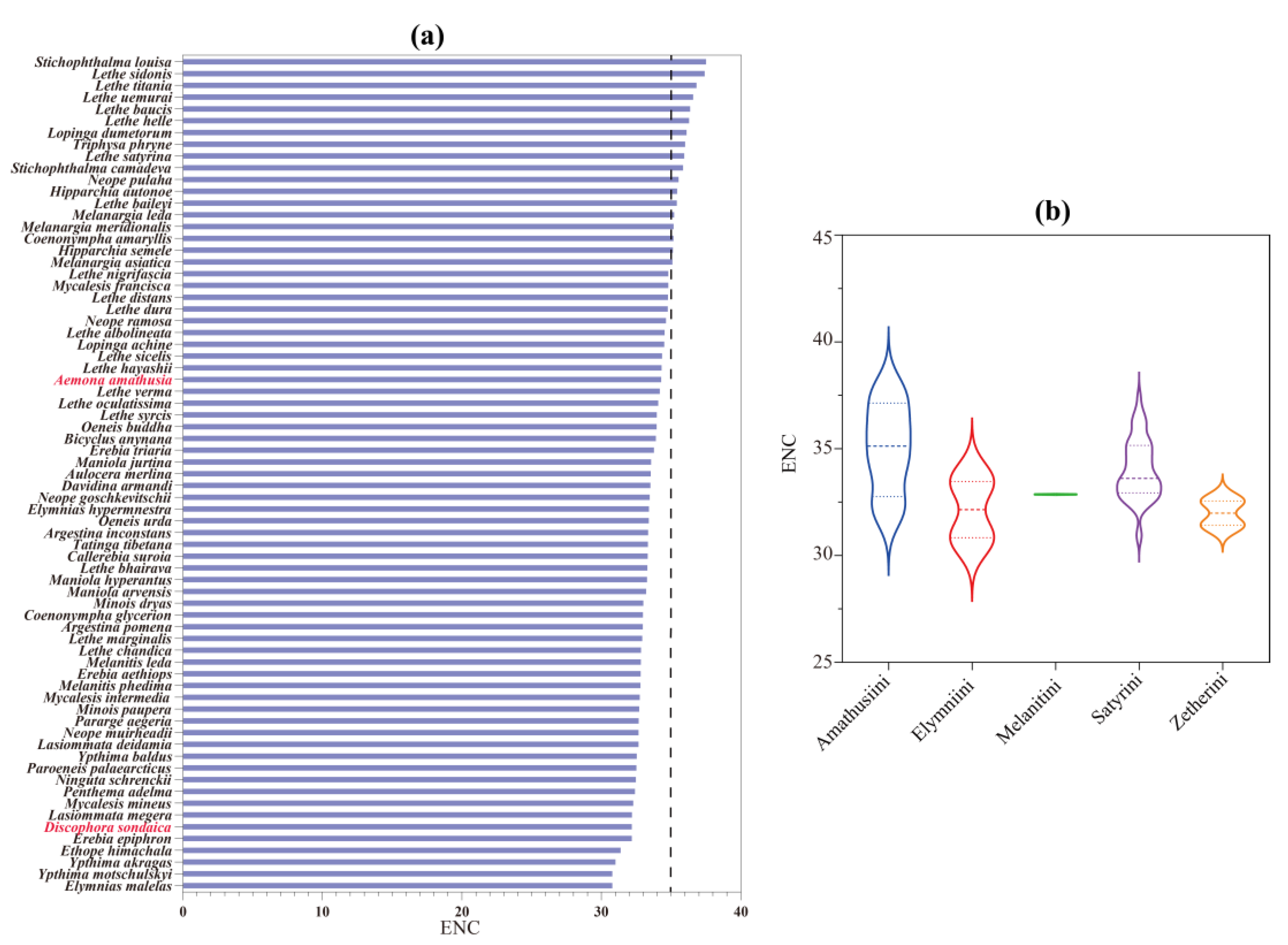
3.2. Protein-Coding Genes and Codon Usage
3.3. Transfer RNA, Ribosomal RNA Genes and Control Region
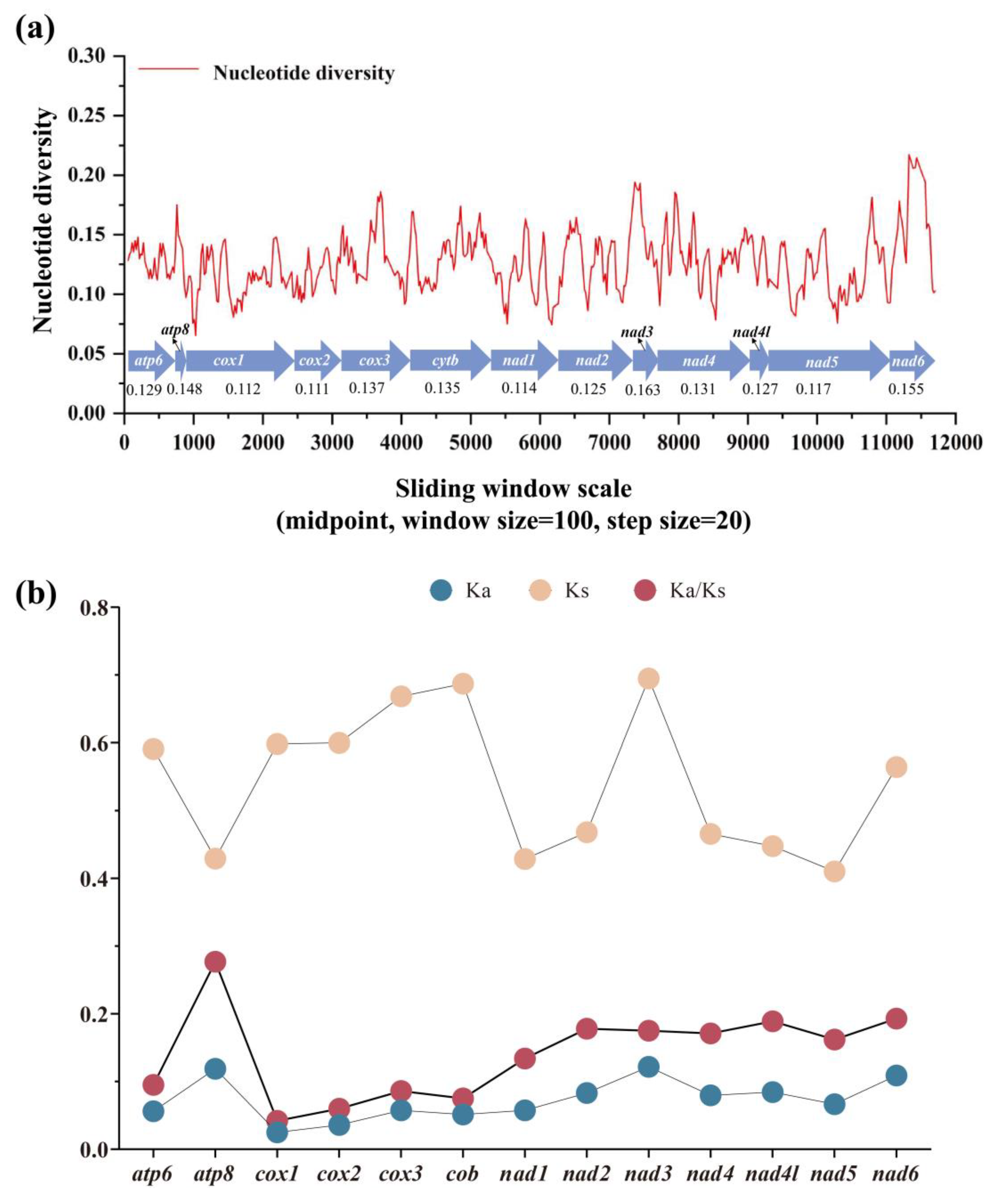
3.4. Nucleotide Diversity and Evolutionary Rate Among Satyrinae Mitogenomes
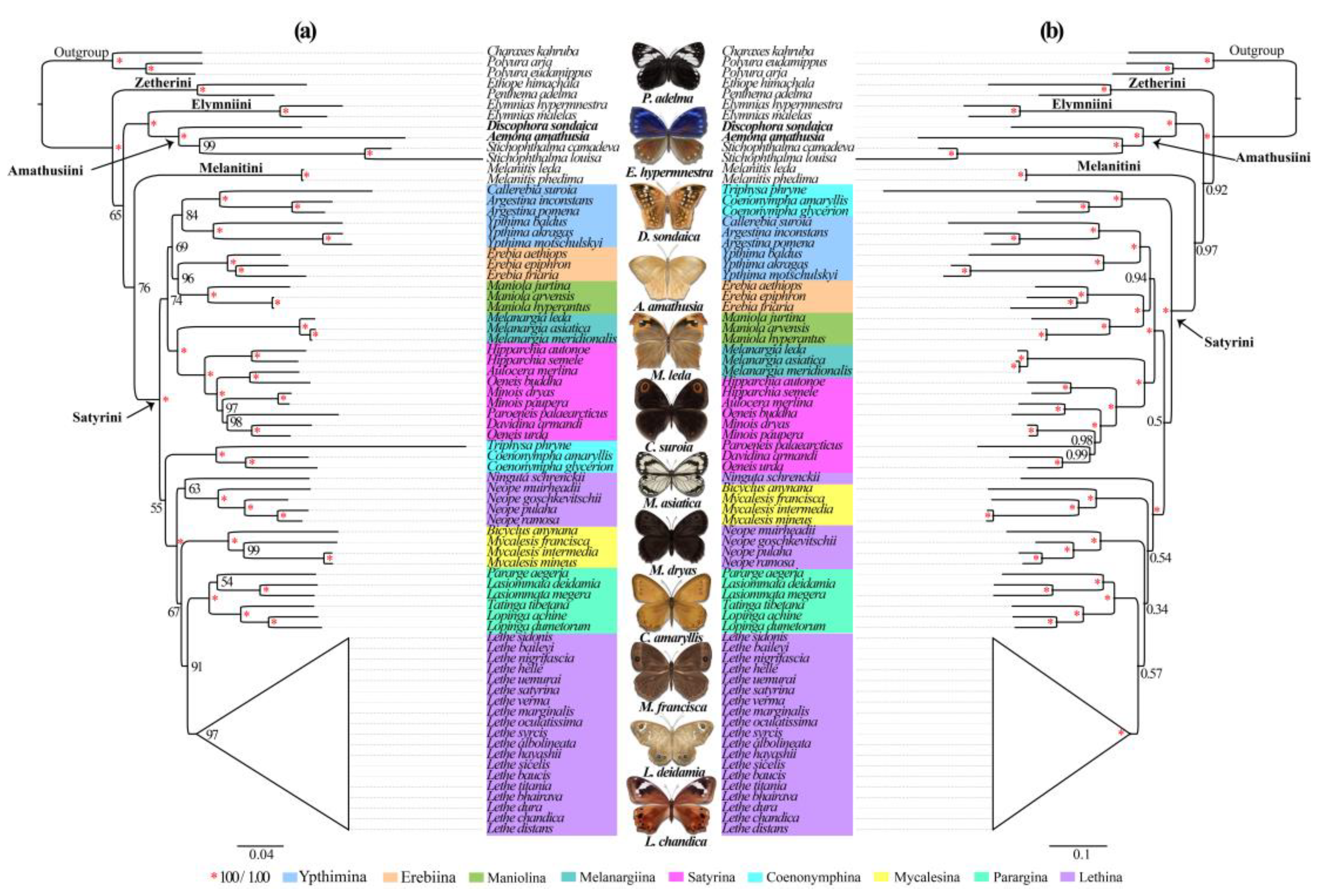
3.5. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, K.Y.; Lee, S.Y.; Bang, I.C.; Nam, Y.K. Complete mitogenome sequence of an endangered freshwater fish, Iksookimia choii (Teleostei; Cypriniformes; Cobitidae). Mitochondrial DNA 2008, 19, 438–445. [Google Scholar] [PubMed]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar]
- Wolstenholme, D.R. Animal Mitochondrial DNA: Structure and Evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [PubMed]
- Zhang, D.X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionarystudies. Biochem. Syst. Ecol. Evol. 1997, 25, 99–120. [Google Scholar]
- Ackery, P.R.; de Jong, R.; Vane-Wright, R.I. The Butterflies: Hedyloidea, Hesperioidea and Papilionoidea. In Handbook of Zoology; Vol. IV Arthropoda: Insecta. Lepidoptera, Moths and Butterflies, vol. 1: Evolution, Kristensen, N.P., Eds.; Walter de Gruyter: Berlin, Germany, 1999; Volume 4, pp. 263–300. [Google Scholar]
- Peña, C.; Wahlberg, N. Prehistorical climate change increased diversification of a group of butterflies. Biol. Letters 2008, 4, 274–278. [Google Scholar]
- Chou, I. Classification and Identification of Chinese Butterflies. Henan Scientific and Technological Publishing House: Zhengzhou, China, 1998.
- Schmitt, T.; Haubrich, K. The genetic structure of the mountain forest butterfly Erebia euryale unravels the late Pleistocene and postglacial history of the mountain coniferous forest biome in Europe. Mol. Ecol. 2008, 17, 2194–2207. [Google Scholar]
- Oliver, J.C.; Tong, X.L.; Gall, L.F.; Piel, W.H.; Monteiro, A. A single origin for nymphalid butterfly eyespots followed by wide spread loss of associated gene expression. PLoS Genet. 2012, 8, e1002893. [Google Scholar]
- Slamova, I.; Klecka, J.; Konvicka, M. Woodland and grassland mosaic from a butterfly perspective: Habitat use by Erebia aethiops (Lepidoptera: Satyridae). Insect Conserv. Diver. 2013, 6, 243–254. [Google Scholar]
- Miller, L.D. The higher classification, phylogeny and zoogeography of the Satyridae (Lepidoptera). Memoirs Am. Entomol. Soc. 1968, 24, 1–174. [Google Scholar]
- Harvey, D.J. Higher classification of the Nymphalidae, Appendix B. In The Development and Evolution of B. In The Development and Evolution of Butterfly Wing Patterns; Nijhout, H.F., Ed.; Smithsonian Institution Press: Washington, DC, USA, 1991; pp. 255–273. [Google Scholar]
- Brower, A.V.Z. Phylogenetic relationships among the Nymphalidae (Lepidoptera) inferred from partial sequences of the wingless gene. P. Roy. Soc. B: Biol. Sci. 2000, 267, 1201–1211. [Google Scholar]
- Freitas, A.; Brown, K. Phylogeny of the Nymphalidae (Lepidoptera). Syst. Biol. 2004, 53, 363–383. [Google Scholar] [CrossRef] [PubMed]
- Wahlberg, N.; Weingartner, E.; Nylin, S. Towards a better understanding of the higher systematics of Nymphalidae (Lepidoptera: Papilionoidea). Mol. Phylogenet. Evol. 2003, 28, 473–484. [Google Scholar] [CrossRef]
- Wahlberg, N.; Braby, M.F.; Brower, A.V.Z. , de Jong, R.; Lee, M.M.; Nylin, S.; Pierce, N.E.; Sperling, F.A.H.; Vila, R.; Warren, A.D.; et al. Synergistic effects of combining morphological and molecular data in resolving the phylogeny of butterflies and skippers. Proc. R. Soc. B. 2005, 272, 1577–1586. [Google Scholar] [CrossRef]
- Peña, C.; Wahlberg, N.; Weingartner, E.; Kodandaramaiah, U.; Brower, A.V.Z. Higher level phylogeny of Satyrinae butterflies (Lepidoptera: Nymphalidae) based on DNA sequence data. Mol. Phylogenet. Evol. 2006, 40, 29–49. [Google Scholar] [CrossRef]
- Wahlberg, N.; Leneveu, J.; Kodandaramaiah, U.; Peña, C.; Nylin, S.; Freitas, A.V.; Brower, A.V. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc. Biol. Sci. 2009, 276, 4295–4302. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.H.; Sun, X.Y.; Wang, Y.L.; Hao, J.S.; Yang, Q. Morphological characters are compatible with mitogenomic data in resolving the phylogeny of nymphalid butterflies (Lepidoptera: Papilionoidea: Nymphalidae). PLoS ONE 2015, 10, e0124349. [Google Scholar] [CrossRef] [PubMed]
- Marín, M.A.; Peña, C.; Freitas, A.V.; Wahlberg, N.; Uribe, S.I. From the phylogeny of the Satyrinae butterflies to the systematics of Euptychiina (Lepidoptera: Nymphalidae): history, progress and prospects. Neotrop. Entomol. 2011, 40, 1–13. [Google Scholar] [CrossRef]
- Yang, M.S.; Zhang, Y.L. Phylogenetic utility of ribosomal genes for reconstructing the phylogeny of five Chinese satyrine tribes (Lepidoptera, Nymphalidae). ZooKeys 2015, 488, 105–120. [Google Scholar]
- Yang, M.S.; Zhang, Y.L. Molecular phylogeny of the butterfly tribe Satyrini (Nymphalidae: Satyrinae) with emphasis on the utility of ribosomal genes mitochondrial 16s rDNA and nuclear 28s rDNA. Zootaxa 2015, 3985, 125–141. [Google Scholar] [CrossRef]
- Peña, C.; Nylin, S.; Wahlberg, N. The radiation of Satyrini butterflies (Nymphalidae: Satyrinae): a challenge for phylogenetic methods. Zool. J. Linn. Soc-lond. 2011, 161, 64–87. [Google Scholar] [CrossRef]
- Yang, M.S.; Song, L.; Zhou, L.; Shi, Y.X.; Song, N.; Zhang, Y.L. Mitochondrial genomes of four satyrine butterflies and phylogenetic relationships of the family Nymphalidae (Lepidoptera: Papilionoidea). Int. J. Biol. Macromol. 2020, 145, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wahlberg, N.; Liao, C.Q.; Wang, C.B.; Ma, F.Z.; Huang, G.H. Fourteen complete mitochondrial genomes of butterflies from the genus Lethe (Lepidoptera, Nymphalidae, Satyrinae) with mitogenome-based phylogenetic analysis. Genomics 2020, 112, 4435–4441. [Google Scholar] [CrossRef]
- Dan, Z.C.; Duan, L.; Chen, Z.N.; Guan, D.L.; Xu, S.Q. Mitogenomes of three satyrid butterfly species (Nymphalidae: Lepidoptera) and reconstructed phylogeny of Satyrinae. Diversity 2021, 13, 468. [Google Scholar] [CrossRef]
- Wu, J.L.; Bao, T.T.; Sun, G.; Xiao, Y.; Fang, Y.; Shi, Q.H. Complete mitochondrial genome of the Woodland Brown, Lopinga achine Scopoli, 1763 (Nymphalidae: Satyrinae) and its phylogenetic analysis. Mitochondrial DNA B 2022, 7, 747–749. [Google Scholar]
- Hu, W.Q.; Mao, K.S.; Dou, L. Complete mitochondrial genome of the Minois paupera Alpheraky, 1888 (Nymphalidae: Satyrinae) and its phylogenetic analysis. Mitochondrial DNA B 2024, 9, 738–742. [Google Scholar]
- Shi, Q.H.; Xie, J.L.; Wu, J.L.; Chen, S.C.; Sun, G.; Zhang, J.C. Characterization of the complete mitochondrial genome of an endemic species in China, Aulocera merlina (Lepidoptera: Nymphalidae: Satyrinae) and phylogenetic analysis within Satyrinae. Ecol. Evol. 2024, 14, e11355. [Google Scholar]
- Wu, L.W.; Lin, L.H.; Lees, D.C.; Hsu, Y.F. Mitogenomic sequences effectively recover relationships within brush-footed butterflies (Lepidoptera: Nymphalidae). BMC Genomics 2014, 15, 468. [Google Scholar]
- Li, X.D.; Hu, H.W.; Zhang, S.L.; Wang, J.W.; Li, R. Characterization of the complete mitochondrial genome of Ypthima baldus (Lepidoptera: Satyrinae) with phylogenetic analysis. Mitochondrial DNA B 2020, 5, 1019–1020. [Google Scholar]
- Shi, Q.H.; Lin, X.Q.; Ye, X.; Xing, J.H.; Dong, G.W. Characterization of the complete mitochondrial genome of Minois dryas (Lepidoptera: Nymphalidae: Satyrinae) with phylogenetic analysis. Mitochondrial DNA B 2019, 4, 1447–1449. [Google Scholar]
- Zhou, Y.; Liang, Z.Y.; Wang, S.Q.; Zhong, H.H.; Wang, N.; Liang, B. A mitogenomic phylogeny of satyrid butterflies and complete mitochondrial genome of Oeneis urda (Lepidoptera: Nymphalidae: Satyrinae). Mitochondrial DNA B 2020, 5, 1344–1345. [Google Scholar]
- Sun, Y.X.; Chen, C. , Geng, X.X.; Li, J. Complete mitochondrial genome of Lasiommata deidamia and its phylogenetic implication to subfamily Satyrinae (Lepidoptera: Nymphalidae). Mitochondrial DNA B 2021, 6, 2943–2945. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, btu170. [Google Scholar]
- Meng, G.L.; Li, Y.Y.; Yang, C.T.; Liu, S.L. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improvedde novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, gkad326. [Google Scholar]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlic´, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: an integrated and scalable desktop platformfor streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Julio, R.; Albert, F.M.; Juan, C.S.D.; Sara, G.R.; Pablo, L.; Sebastián, E.R.O.; Alejandro, S.G. DnaSP v6: DNA sequence polymor-phism analysis of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar]
- Peden, J.F. Analysis of codon usage. Univ Nottingham. 2000, 90, 73–74. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Phylogenet. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Ranwez, V.; Douzery, E.J.P.; Cambon, C.; Chantret, N.; Delsuc, F. MACSE v2: Toolkit for the alignment of coding sequences accounting for frame shifts and stop codons. Mol. Biol. Evol. 2018, 35, 2582–2584. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics, 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar]
- Rambaut, FigTree (Version 1.4.4). Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 28 December 2024).
- Shi, Q.H.; Zhang, W.; Hao, J.S. The complete mitochondrial genome of Callerebia suroia (Lepidoptera: Nymphalidae: Satyrinae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 1463–1465. [Google Scholar] [CrossRef]
- Kim, M.J.; Wan, X.L.; Kim, K.G.; Hwang, J.S.; Kim, I. Complete nucleotide sequence and organization of the mitogenome of endangered Eumenis autonoe (Lepidoptera: Nymphalidae). Afr. J. Biotechnol. 2010, 9, 735–754. [Google Scholar]
- Teixeira da Costa, L.F. The complete mitochondrial genome of Parage aegeria (Insecta: Lepidoptera: Papilionidae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 551–552. [Google Scholar]
- Liu, G.C.; Chang, Z.; Chen, L.; He, J.W.; Dong, Z.W.; Yang, J.; Lu, S.H.; Zhao, R.P.; Wan, W.T.; Ma, G.L.; et al. Genome size variation in butterflies (Insecta, Lepidoptera, Papilionoidea): a thorough phylogenetic comparison. Syst. Entomol. 2020, 45, 571–582. [Google Scholar]
- Tang, M.; Tan, M.H.; Meng, G.L.; Yang, S.Z.; Su, X.; Liu, S.L.; Song, W.H.; Li, Y.Y.; Wu, Q.; Zhang, A.B.; et al. Multiplex sequencing of pooled mitochondrial genomes—a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 2014, 42, e166. [Google Scholar]
- Wu, Y.P.; Lu, J.J.; Yang, J.; Wang, J.P.; Cao, T.W.; Fan, R.J. Complete mitochondrial genome of Mycalesis intermedia (Lepidoptera: Nymphalidae). Mitochondrial DNA B 2020, 5, 703–704. [Google Scholar]
- Huang, D.Y.; Hao, J.S.; Zhang, W.; Su, T. J.; Wang, Y.; Xu, X. F. The complete mitochondrial genome of Melanargia asiatica (Lepidoptera: Nymphalidae: Satyrinae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 806–808. [Google Scholar]
- Lohse, K.; Weir, J. The genome sequence of the meadow brown, Maniola jurtina (Linnaeus, 1758). Wellcome Open Res. 2021, 6, 296. [Google Scholar]
- Nagata, N.; Tsujimura, I.; Sato, A. The complete mitochondrial genomes of two Japanese endemic Satyrinae butterflies, Neope goschkevitschii and Lethe sicelis (Lepidoptera, Nymphalidae). Mitochondrial DNA B 2020, 5, 2243–2245. [Google Scholar] [CrossRef]
- Fan, C.; Xu, C.; Li, J.L.; Lei, Y.; Gao, Y.; Xu, C.R.; Wang, R.J. Complete mitochondrial genome of a satyrid butterfly, Ninguta schrenkii (Lepidoptera: Nymphalidae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 80–81. [Google Scholar]
- Li, J.L.; Xu, C.; Lei, Y.; Fan, C.; Gao, Y.; Xu, C.R.; Wang, R.J. Complete mitochondrial genome of a satyrid butterfly, Lethe albolineata (Lepidoptera: Nymphalidae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 4195–4196. [Google Scholar]
- Zhou, L.; Yang, C.; Zhai, Q.; Zhang, Y.L. The complete mitochondrial genome sequence of Coenonympha amaryllis and monophyly of Satyrinae (Lepidoptera: Nymphalidae). Mitochondrial DNA B 2020, 5, 1223–1224. [Google Scholar]
- Zhang, W.; Gan, S.S.; Zuo, N.; Chen, C.H.; Wang, Y.; Hao, J.S. The complete mitochondrial genome of Triphysa phryne (Lepidoptera: Nymphalidae: Satyrinae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 474–475. [Google Scholar]
- Lohse, O.; Lohse, K. The genome sequence of the scotch argus butterfly, Erebia aethiops (Esper, 1777). Wellcome Open Res. 2022, 7, 217. [Google Scholar]
- Shi, Q.H.; Zhao, F.; Hao, J.S.; Yang, Q. Complete mitochondrial genome of the Common Evening Brown, Melanitis leda Linnaeus (Lepidoptera: Nymphalidae: Satyrinae). Mitochondrial DNA, 2013, 24, 492–494. [Google Scholar]
- Wei, S.J.; Shi, M.; Chen, X.X.; Sharkey, M.J.; van Achterberg, C.; Ye, G.Y.; He, J.H. New views on strand asymmetry in insect mitochondrial genomes. PLoS One 2010, 5, e12708. [Google Scholar]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar]
- Lu, H.F.; Su, T.J.; Luo, A.R.; Zhu, C.D.; Wu, C.S. Characterization of the complete mitochondrion genome of Diurnal moth Amata emma (Butler) (Lepidoptera: Erebidae) its phylogenetic implications. PLoS ONE 2013, 8, e72410. [Google Scholar]
- Hershberg, R.; Petrov, D.A. Selection on codon bias. Annu. Rev. Genet. 2008, 42, 287–299. [Google Scholar] [PubMed]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar]
- Wright, D. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar]
- Salvato, P.; Simonato, M.; Battisti, A.; Negrisolo, E. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genomics 2008, 9, 331. [Google Scholar]
- Chen, S.; Li, F.H.; Lan, X.E.; You, P. The complete mitochondrial genome of Pycnarmon lactiferalis (Lepidoptera: Crambidae). Mitochondrial DNA B 2016, 1, 638–639. [Google Scholar]
- Chen, Z.T.; Du, Y.Z. The first two mitochondrial genomes from Taeniopterygidae (Insecta: Plecoptera): structural features and phylogenetic implications. Int. J. Biol. Macromol. 2017, 111, 70–76. [Google Scholar]
- Yang, M.S.; Song, L.; Shi, Y.X.; Li, J.H.; Zhang, Y.L.; Song, N. The first mitochondrial genome of the family Epicopeiidae and higher-level phylogeny of Macroheterocera (Lepidoptera: Ditrysia). Int. J. Biol. Macromol. 2019, 136, 123–132. [Google Scholar]
- Vila, M.; Björklund, M. The utility of the neglected mitochondrial control region for evolutionary studies in Lepidoptera (Insecta). J. Mol. Evol. 2004, 58, 280–290. [Google Scholar] [CrossRef]
- Jia, W.Z.; Yan, H.B.; Guo, A.J.; Zhu, X.Q.; Wang, Y.C.; Shi, W.G.; Chen, H.T.; Zhan, F.; Zhang, S.H.; Fu, B.Q.; Littlewood, D.T.J.; Cai, X.P. Complete mitochondrial genomes of Taenia multiceps, T. hydatigena and T. pisiformis: Additional molecular markers for a tapeworm genus of human and animal health significance. BMC Genomics, 2010, 11, 447. [Google Scholar]
- Ma, L.Y.; Liu, F.F.; Chiba, H.; Yuan, X.Q. The mitochondrial genomes of three skippers: insights into the evolution of the family Hesperiidae (lepidoptera). Genomics 2020, 112, 432–441. [Google Scholar]
- Yan, Z.T.; Tang, X.Y.; Yang, D.; Fan, Z.H.; Luo, S.T.; Chen, B. Phylogenetic and comparative genomics study of Papilionidae based on mitochondrial genomes. Genes 2024, 15, 964. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Fang, L.; Zhang, Y. The complete mitochondrial genomes of four species in the subfamily Limenitidinae (Lepidoptera, Nymphalidae) and a phylogenetic analysis. Insects 2022, 13, 16. [Google Scholar]
- Meiklejohn, C.D.; Montooth, K.L.; Rand, D.M. Positive and negative selection on the mitochondrial genome. Trends Genet. 2007, 23, 259–263. [Google Scholar] [PubMed]
- Sun, Y.X.; Chen, C.; Geng, X.X.; Li, J. Complete mitochondrial genome of Lasiommata deidamia and its phylogenetic implication to subfamily Satyrinae (Lepidoptera: Nymphalidae). Mitochondrial DNA B 2021, 6, 2943–2945. [Google Scholar]
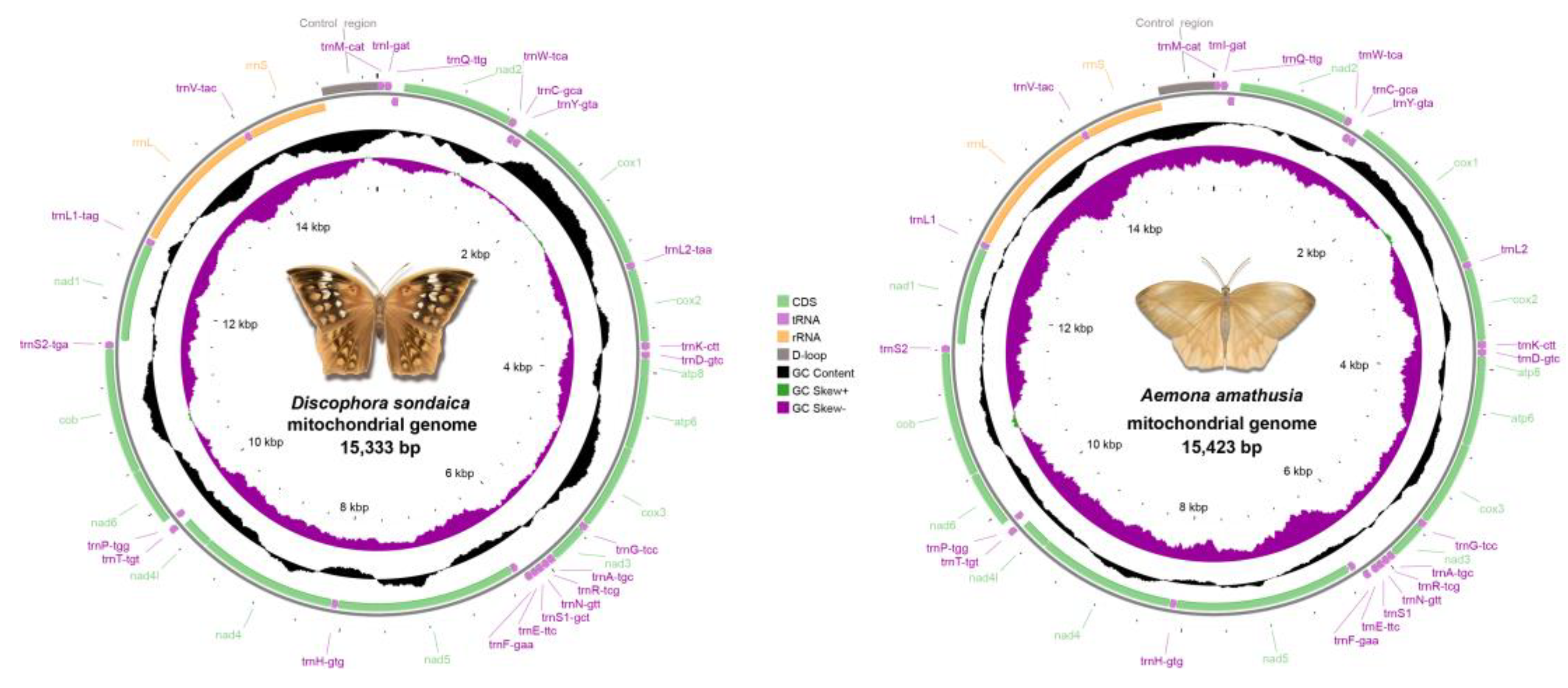
| Gene | Coding strand | Position | Size (bp) | IGN | Start codon | Stop codon | |||||
| DS | AA | DS | AA | DS | AA | DS | AA | DS | AA | ||
| trnM | J | 1-69 | 1-66 | 69 | 66 | 0 | 0 | ||||
| trnI | J | 70-136 | 69-133 | 67 | 65 | 0 | 2 | ||||
| trnQ | N | 134-201 | 131-199 | 68 | 69 | -3 | -3 | ||||
| nad2 | J | 248-1261 | 245-1258 | 1014 | 1014 | 46 | 45 | ATT | ATC | TAA | TAA |
| trnW | J | 1260-1327 | 1257-1323 | 68 | 67 | -2 | -2 | ||||
| trnC | N | 1320-1383 | 1316-1383 | 64 | 68 | -8 | -8 | ||||
| trnY | N | 1384-1450 | 1384-1449 | 67 | 66 | 0 | 0 | ||||
| cox1 | J | 1453-2983 | 1471-3001 | 1531 | 1531 | 2 | 21 | CGA | CGA | T | T |
| trnL2 | J | 2984-3051 | 3002-3068 | 68 | 67 | 0 | 0 | ||||
| cox2 | J | 3052-3727 | 3070-3742 | 676 | 673 | 0 | 1 | ATG | ATG | T | T |
| trnK | J | 3728-3798 | 3743-3813 | 71 | 71 | 0 | 0 | ||||
| trnD | J | 3810-3874 | 3816-3880 | 65 | 65 | 11 | 2 | ||||
| atp8 | J | 3875-4039 | 3881-4039 | 165 | 159 | 0 | 0 | ATC | ATG | TAA | TAA |
| atp6 | J | 4033-4710 | 4033-4710 | 678 | 678 | -7 | -7 | ATG | ATG | TAA | TAA |
| cox3 | J | 4710-5498 | 4710-5498 | 789 | 789 | -1 | -1 | ATG | ATG | TAA | TAA |
| trnG | J | 5501-5567 | 5501-5566 | 67 | 66 | 2 | 2 | ||||
| nad3 | J | 5568-5915 | 5564-5914 | 348 | 351 | 0 | -3 | ATT | ATA | TAA | TAA |
| trnA | J | 5919-5988 | 5916-5979 | 70 | 64 | 3 | 1 | ||||
| trnR | J | 5988-6049 | 5979-6040 | 62 | 62 | -1 | -1 | ||||
| trnN | J | 6051-6119 | 6041-6106 | 69 | 66 | 1 | 0 | ||||
| trnS1 | J | 6117-6177 | 6104-6164 | 61 | 61 | -3 | -3 | ||||
| trnE | J | 6178-6242 | 6194-6257 | 65 | 64 | 0 | 29 | ||||
| trnF | N | 6241-6306 | 6262-6327 | 66 | 66 | -2 | 4 | ||||
| nad5 | N | 6307-8044 | 6333-8072 | 1738 | 1740 | 0 | 5 | ATT | ATT | T | TAA |
| trnH | N | 8045-8108 | 8073-8137 | 64 | 65 | 0 | 0 | ||||
| nad4 | N | 8109-9447 | 8138-9476 | 1339 | 1339 | 0 | 0 | ATG | ATG | T | T |
| nad4l | N | 9449-9739 | 9477-9764 | 291 | 288 | 1 | 0 | ATG | ATG | TAA | TAA |
| trnT | J | 9742-9806 | 9767-9830 | 65 | 64 | 2 | 2 | ||||
| trnP | N | 9807-9871 | 9831-9895 | 65 | 65 | 0 | 0 | ||||
| nad6 | J | 9874-10404 | 9901-10428 | 531 | 528 | 2 | 5 | ATT | ATA | TAA | TAA |
| cob | J | 10404-11556 | 10432-11586 | 1153 | 1155 | -1 | 3 | ATG | ATG | T | TAA |
| trnS2 | J | 11557-11622 | 11586-11650 | 66 | 65 | 0 | -1 | ||||
| nad1 | N | 11625-12579 | 11656-12609 | 955 | 954 | 2 | 5 | ATG | ATA | T | TAA |
| trnL1 | N | 12580-12648 | 12613-12681 | 69 | 69 | 0 | 3 | ||||
| rrnL | N | 12663-13993 | 12684-14082 | 1331 | 1399 | 14 | 2 | ||||
| trnV | N | 13994-14057 | 14083-14148 | 64 | 66 | 0 | 0 | ||||
| rrnS | N | 14058-14827 | 14149-14915 | 770 | 767 | 0 | 0 | ||||
| Control region | 14828-15333 | 14916-15423 | 506 | 508 | 0 | 0 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




