Submitted:
21 March 2025
Posted:
24 March 2025
You are already at the latest version
Abstract
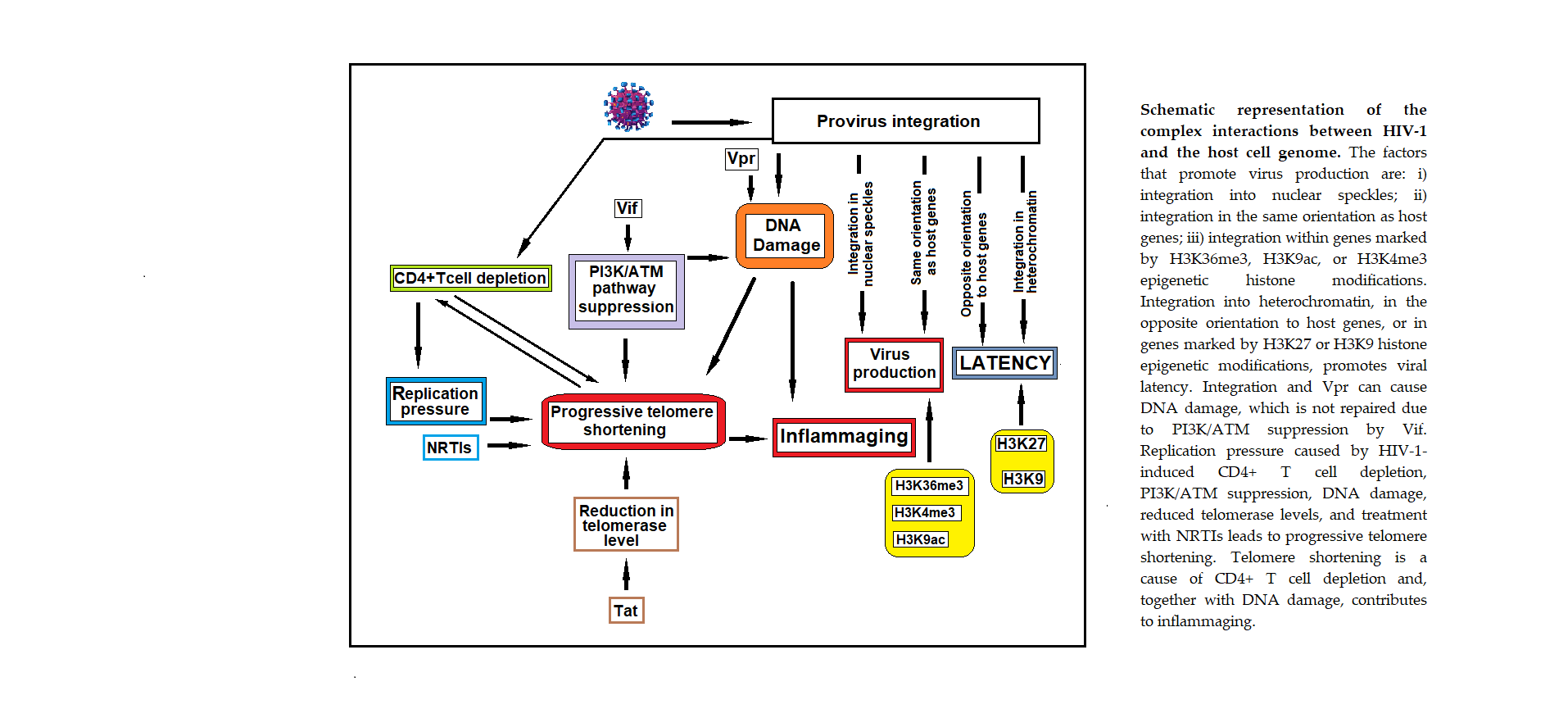
Keywords:
1. Introduction
2. Integration and Post-Integration Stages in HIV-1 Infection
2.1. HIV-1 Provirus Integration
2.2. Post-Integrational DNA Repair
2.3. HIV-1 Provirus Integration Sites
2.4. HIV-1 Provirus Orientation
3. HIV-1-Induced Host Genome Damage
3.1. HIV-1-Induced DNA Breaks
3.2. DNA Damage and CD4+ T Cell Depletion
3.3. DNA Damage and Its Role in Inflammaging in HIV-1 Infected Individuals
4. HIV-1 Integration and CD4+ T Cell Clonal Expansion
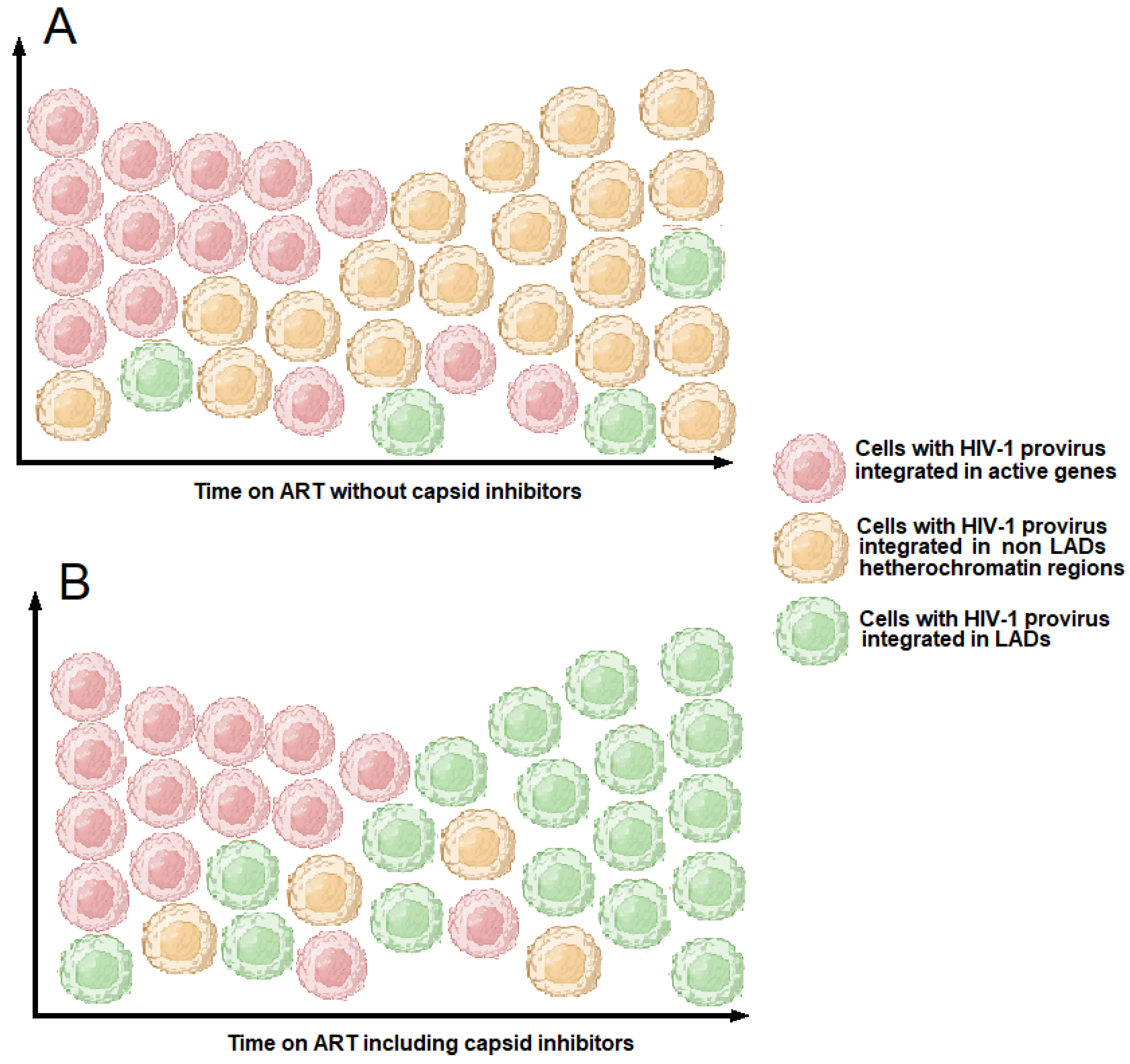
5. Capsid and Its Role in Driving HIV-1 Integration into Gene-Dense Regions or in Lamina-Associated Domains (LADs)
6. Comorbidities Caused by HIV-1-Induced DNA Damage
6.1. Cancer and Provirus Integration
6.2. Pulmonary Arterial Hypertension
6.3. DNA Damage in Renal Tubule Epithelial Cells
6.4. Neurological Dysfunctions and DNA Damage
7. New Therapeutic Strategies to Treat HIV Infection
7.1. HIV-1 Treatment Based on Capsid Inhibitors
7.2. The "Shock and Kill" Therapeutic Approach
7.3. HIV-1 Treatment Based on Silencing or Knocking out the Provirus
8. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Krebs, A.S.; Mendonça, L.M.; Zhang, P. Structural Analysis of Retrovirus Assembly and Maturation. Viruses 2021, 14, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Maertens, G.N.; Engelman, A.N.; Cherepanov, P. Structure and function of retroviral integrase. Nat. Rev. Microbiol. 2022, 20, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Grawenhoff, J.; Engelman, A.N. Retroviral integrase protein and intasome nucleoprotein complex structures. World J. Biol. Chem. 2017, 8, 32–44. [Google Scholar] [CrossRef]
- Dopkins, N.; O’Mara, M.M.; Lawrence, E.; Fei, T. : Sandoval-Motta, S.; Nixon, D.F.; Bendall, M.L. A field guide to endogenous retrovirus regulatory networks. Mol. Cell. 2022, 82, 3763–3768. [Google Scholar] [CrossRef]
- Zahn, J.; Kaplan, M.H.; Fischer, S.; Dai, M.; Meng, F.; Saha, A.K.; Cervantes, P.; Chan, S.M.; Dube, D.; Omenn, G.S.; et al. Expansion of a novel endogenous retrovirus throughout the pericentromeres of modern humans. Genome Biol. 2015, 16, 74. [Google Scholar] [CrossRef]
- Roshal, M.; Kim, B.; Zhu, Y.; Nghiem, P.; Planelles, V. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J. Biol. Chem. 2003, 278, 25879–86. [Google Scholar] [CrossRef]
- Zimmerman, E.S.; Sherman, M.P.; Blackett, J.L.; Neidleman, J.A.; Kreis, C.; Mundt, P.; Williams, S.A.; Warmerdam, M.; Kahn, J.; Hecht, F.M.; et al. Human immunodeficiency virus type 1 Vpr induces DNA replication stress in vitro and in vivo. J. Virol. 2006, 80, 10407–18. [Google Scholar] [CrossRef]
- Iijima, K.; Kobayashi, J.; Ishizaka, Y. Structural alteration of DNA induced by viral protein R of HIV-1 triggers the DNA damage response. Retrovirology 2018, 15, 8. [Google Scholar] [CrossRef]
- Li, D.; Lopez, A.; Sandoval, C.; Nichols, D.R.; Fregoso, O.I. HIV Vpr modulates the host DNA damage response at two independent steps to damage DNA and repress DSB repair. MBio 2020, 11, e00940–20. [Google Scholar] [CrossRef]
- Wong, T.; Luperchio, A.M.; Riley, S.; Salamango, D.J. Inhibition of ATM-directed antiviral responses by HIV-1 Vif. PLoS Pathog. 2023, 19, e1011634. [Google Scholar] [CrossRef]
- Wagner, T.A.; McLaughlin, S.; Garg, K.; Cheung, C.Y.; Larsen, B.B.; Styrchak, S.; Huang, H.C.; Edlefsen, P.T.; Mullins, J.I.; Frenkel, L.M. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 2014, 345, 570–573. [Google Scholar] [CrossRef]
- Dragoni, F.; Kwaa, A.K.; Traut, C.C.; Veenhuis, R.T.; Woldemeskel, B.A.; Camilo-Contreras, A.; Raymond, H.E.; Dykema, A.G.; Scully, E.P.; Rosecrans, A.M.; et al. Proviral location affects cognate peptide-induced virus production and immune recognition of HIV-1-infected T cell clones. J. Clin. Investig. 2023, 133, e171097. [Google Scholar] [CrossRef]
- Zhao, J.; Nguyen, L.N.T.; Dang, X.; Cao, D.; Khanal, S.; Schank, M.; Thakuri, B.K.C.; Ogbu, S.C.; Morrison, Z.D.; Wu, X.Y. , et al. ATM deficiency accelerates DNA damage, telomere erosion, and premature T cell aging in HIV-infected individuals on antiretroviral therapy. Front. Immunol. 2019, 10, 2531. [Google Scholar] [CrossRef]
- Khanal, S.; Tang, Q.; Cao, D.; Zhao, J.; Nguyen, L.N.; Oyedeji, O.S.; Dang, X.; Nguyen, L.N.T.; Schank, M.; Thakuri, B.K.C.; et al. Telomere and ATM Dynamics in CD4 T-Cell Depletion in Active and Virus-Suppressed HIV Infections. J. Virol. 2020, 94, e01061–20. [Google Scholar] [CrossRef]
- Simenauer, A.; Nozik-Grayck, E.; Cota-Gomez, A. The DNA Damage Response and HIV-Associated Pulmonary Arterial Hypertension. Int. J. Mol. Sci. 2020, 21, 3305. [Google Scholar] [CrossRef]
- Wang, P.; Ouyang, J.; Jia, Z.; Zhang, A.; Yang, Y. Roles of DNA damage in renal tubular epithelial cells injury. Front. Physiol. 2023, 14, 1162546. [Google Scholar] [CrossRef]
- Delint-Ramirez, I.; Madabhushi, R. NPAS4 juggles neuronal activity-dependent transcription and DSB repair with NuA4. Mol. Cell. 2023, 83, 1208–1209. [Google Scholar] [CrossRef]
- Chen, B. Molecular mechanism of HIV-1 entry. Trends Microbiol. 2019, 27, 878–891. [Google Scholar] [CrossRef]
- Novikova, M.; Zhang, Y.; Freed, E.O.; Peng, K. Multiple roles of HIV-1 capsid during the virus replication cycle. Virol. Sin. 2019, 34, 119–134. [Google Scholar] [CrossRef]
- Arhel, N. Revisiting HIV-1 uncoating. Retrovirology 2010, 7, 96–105. [Google Scholar] [CrossRef]
- Bukrinsky, M.I.; Haggerty, S.; Dempsey, M.P.; Sharova, N.; Adzhubel, A.; Spitz, L.; Lewis, P.; Goldfarb, D.; Emerman, M.; Stevenson, M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 1993, 365, 666–9. [Google Scholar] [CrossRef] [PubMed]
- Depienne, C.; Mousnier, A.; Leh, H.; Le Rouzic, E.; Dormont, D.; Benichou, S.; Dargemont, C. Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 2001, 276, 18102–18107. [Google Scholar] [CrossRef] [PubMed]
- Gallay, P.; Hope, T.; Chin, D.; Trono, D. HIV-1 infection of nondividing cells through the recognition of Integrase by the Importin/Karyopherin pathway. Proc. Natl. Acad. Sci. USA 1997, 94, 9825–9830. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.L.; Spearman, P.; Ratner, L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J. Virol. 1993, 67, 542–6550. [Google Scholar] [CrossRef]
- Yamashita, M.; Emerman, M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 2004, 78, 5670–5678. [Google Scholar] [CrossRef]
- Yamashita, M.; Perez, O.; Hope, T.J.; Emerman, M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 2007, 3, 1502–1510. [Google Scholar] [CrossRef]
- Dwivedi, R.; Prakash, P.; Kumbhar, B.V.; Balasubramaniam, M.; Dash, C. HIV-1 capsid and viral DNA integration. mBio 2024, 15, e0021222. [Google Scholar] [CrossRef]
- Burdick, R.C.; Morse, M.; Rouzina, I.; Williams, M.C.; Hu, W.S.; Pathak, V.K. HIV-1 uncoating requires long double-stranded reverse transcription products. Sci. Adv. 2024, 10, eadn7033. [Google Scholar] [CrossRef]
- Lesbats, P.; Engelman, A. N.; Cherepanov, P. Retroviral DNA integration. Chem. Rev. 2016, 116, 12730–12757. [Google Scholar]
- Anisenko, A.; Nefedova, A.; Agapkina, Y.; Gottikh, M. Both ATM and DNA-PK Are the Main Regulators of HIV-1 Post-Integrational DNA Repair. Int. J. Mol. Sci. 2023, 24, 2797–2809. [Google Scholar] [CrossRef]
- Vincent, K. A.; York-Higgins, D.; Quiroga, M.; Brown, P. O. Host sequences flanking the HIV provirus. Nucleic Acids Res. 1990, 18, 6045–6047. [Google Scholar] [CrossRef] [PubMed]
- Vink, C.; Groenink, M.; Elgersma, Y.; Fouchier, R. A.; Tersmette, M.; Plasterk, R. H. Analysis of the junctions between human immunodeficiency virus type 1 proviral DNA and human DNA. J. Virol. 1990, 64, 5626–5627. [Google Scholar] [CrossRef] [PubMed]
- Anisenko, A.N.; Knyazhanskaya, E.S.; Isaguliants, M.G.; Gottikh, M.B. A qPCR assay for measuring the post-integrational DNA repair in HIV-1 replication. J. Virol. Methods. 2018, 262, 12–19. [Google Scholar] [CrossRef]
- Anisenko, A.N.; Nefedova, A.A.; Kireev, I.I.; Gottikh, M.B. Post-Integrational DNA Repair of HIV-1 Is Associated with Activation of the DNA-PK and ATM Cellular Protein Kinases and Phosphorylation of Their Targets. Biochemistry (Mosc) 2024, 89, 1122–1132. [Google Scholar] [CrossRef]
- Barnes, G.; Rio, D. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1997, 94, 867–72. [Google Scholar] [CrossRef]
- Schroder, A.R.; Shinn, P.; Chen, H.; Berry, C.; Ecker, J.R.; Bushman, F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 2002, 110, 521–529. [Google Scholar] [CrossRef]
- Wang, G.P.; Ciuffi, A.; Leipzig, J.; Berry, C.C.; Bushman, F.D. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007, 17, 1186–1194. [Google Scholar] [CrossRef]
- Roth, S.L.; Malani, N.; Bushman, F.D. Gammaretroviral integration into nucleosomal target DNA in vivo. J. Virol. 2011, 85, 7393–7401. [Google Scholar] [CrossRef]
- Sowd, G.A.; Serrao, E.; Wang, H.; Wang, W.; Fadel, H.J.; Poeschla, E.M.; Engelman, A.N. A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 2016, 113, E1054–E1063. [Google Scholar] [CrossRef]
- Kvaratskhelia, M.; Sharma, A.; Larue, R.C.; Serrao, E.; Engelman, A. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res. 2014, 42, 10209–10225. [Google Scholar] [CrossRef]
- Cohn, L.B.; Silva, I.T.; Oliveira, T.Y.; Rosales, R.A.; Parrish, E.H.; Learn, G.H.; Hahn, B.H.; Czartoski, J.L.; McElrath, M.J.; Lehmann, C.; et al. HIV-1 integration landscape during latent and active infection. Cell 2015, 160, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Maldarelli, F.; Wu, X.; Su, L.; Simonetti, F.R.; Shao, W.; Hill, S.; Spindler, J.; Ferris, A.L.; Mellors, J.W.; Kearney, M.F.; et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 2014, 345, 179–183. [Google Scholar] [CrossRef]
- Saleh, S.; Lu, H.K.; Evans, V.; Harisson, D.; Zhou, J.; Jaworowski, A.; Sallmann, G.; Cheong, K.Y.; Mota, T.M.; Tennakoon, S.; et al. HIV integration and the establishment of latency in CCL19-treated resting CD4+ T cells require activation of NF-κB. Retrovirology 2016, 13, 49–60. [Google Scholar] [CrossRef]
- Brady, T.; Agosto, L.M.; Malani, N.; Berry, C.C.; O’Doherty, U.; Bushman, F. HIV integration site distributions in resting and activated CD4 + T cells infected in culture. AIDS 2009, 23, 1461–1471. [Google Scholar] [CrossRef]
- Engelman, A.N.; Maertens, G.N. Virus-host interactions in retrovirus integration. In Retrovirus-Cell Interactions; Parent, L.J., Ed.; Academic Press: San Diego, CA, USA, 2018; pp. 163–198. [Google Scholar]
- Bedwell, G.J.; Engelman, A.N. Factors that mold the nuclear landscape of HIV-1 integration. Nucleic Acids Res. 2021, 49, 621–635. [Google Scholar] [CrossRef]
- Singh, P.K.; Bedwell, G.J.; Engelman, A.N. Spatial and genomic correlates of HIV-1 integration site targeting. Cells 2022, 11, 655. [Google Scholar] [CrossRef]
- Yedavalli, V.R.K.; Jeang, K.T. Methylation: a regulator of HIV-1-1 replication? Retrovirology.
- Willemsen, N.M.; Hitchen, E.M.; Bodetti, T.J.; Apolloni, A.; Warrilow, D.; Piller, S.C.; Harrich, D. Protein methylation is required to maintain optimal HIV-1-1 infectivity. Retrovirology 2006, 3, 92. [Google Scholar] [CrossRef]
- Sapp, N.; Burge, N.; Cox, K.; Prakash, P.; Balasubramaniam, M.; Thapa, S.; Christensen, D.; Li, M.; Linderberger, J.; Kvaratskhelia, M.; et al. HIV-1 preintegration complex preferentially integrates the viral DNA into nucleosomes containing trimethylated histone 3-lysine 36 modification and flanking linker DNA. J. Virol. 2022, 96, e01011–22. [Google Scholar] [CrossRef]
- Vansant, G.; Chen, H.C.; Zorita, E.; Trejbalová, K.; Miklík, D.; Filion, G.; Debyser, Z. The chromatin landscape at the HIV-1 provirus integration site determines viral expression. Nucleic Acids Res. 2020, 48, 7801–7817. [Google Scholar] [CrossRef]
- Bussey-Sutton, C.R.; Ward, A.; Fox, J.A. Turner, A.W.; Peterson, J.J.; Emery, A.; Longoria, A.R.; Gomez-Martinez, I.; Jones, C.; Hepperla, A.; et al. The histone methyltransferase SETD2 regulates HIV expression and latency. PLoS Pathog. 2024, 20, e1012281. [Google Scholar]
- Nguyen, K.; Karn, J. The sounds of silencing: dynamic epigenetic control of HIV latency. Curr. Opin. HIV AIDS. 2024, 19, 102–109. [Google Scholar] [CrossRef]
- Nikolaitchik, O.A.; Islam, S.; Kitzrow, J.P.; Duchon, A.; Cheng, Z.; Liu, Y.; Rawson, J.M.O.; Shao, W.; Nikolaitchik, M.; Kearney, M.F.; et al. HIV-1 usurps transcription start site heterogeneity of host RNA polymerase II to maximize replication fitness. Proc. Natl. Acad. Sci. U S A 2023, 120, e2305103120. [Google Scholar] [CrossRef]
- Ciuffi, A.; Llano, M.; Poeschla, E.; Hoffmann, C.; Leipzig, J.; Shinn, P.; Ecker, J.R.; Bushman, F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005, 11, 1287–1289. [Google Scholar] [CrossRef] [PubMed]
- Shun, M.C.; Raghavendra, N.K.; Vandegraaff, N.; Daigle, J.E.; Hughes, S.; Kellam, P.; Cherepanov, P.; Engelman, A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007, 21, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Plumb, M.R.; Ferris, A.L.; Iben, J.R.; Wu, X.; Fadel, H.J.; Luke, B.T.; Esnault, C.; Poeschla, E.M.; Hughes, S.H.; et al. LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced genes. Genes Dev. 2015, 29, 2287–2297. [Google Scholar] [CrossRef]
- Busschots, K.; Vercammen, J.; Emiliani, S.; Benarous, R.; Engelborghs, Y.; Christ, F.; Debyser, Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 2005, 280, 17841–17847. [Google Scholar] [CrossRef]
- Llano, M.; Delgado, S.; Vanegas, M.; Poeschla, E.M. Lens epithelium-derived growth factor/p75 prevents proteasomal degradation of HIV-1 integrase. J. Biol. Chem. 2004, 279, 55570–55577. [Google Scholar] [CrossRef]
- Cherepanov, P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007, 35, 113–124. [Google Scholar] [CrossRef]
- Kok, Y.L.; Vongrad, V.; Chaudron, S.E.; Shilaih, M.; Leemann, C.; Neumann, K.; Kusejko, K.; Di Giallonardo, F.; Kuster, H.; Braun, D,L. HIV-1 integration sites in CD4+ T cells during primary, chronic, and late presentation of HIV-1 infection. J.C.I. Insight 2021, 6, e143940. [Google Scholar] [CrossRef]
- Miura, Y.; Morooka, M.; Sax, N.; Roychoudhuri, R.; Itoh-Nakadai, A.; Brydun, A.; Funayama, R.; Nakayama, K.; Satomi, S.; Matsumoto, M.; et al. Bach2 promotes B cell receptor-induced proliferation of B lymphocytes and represses cyclin-dependent kinase inhibitors. J. Immunol. 2018, 200, 2882–2893. [Google Scholar] [CrossRef]
- Weng, X.; Zheng, M.; Liu, Y.; Lou, G. The role of Bach2 in regulating CD8 + T cell development and function. Cell. Commun. Signal. 2024, 22, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Shaposhnikov, D.; Kuffer, C.; Storchova, Z.; & Posern, G.; Posern, G. Myocardin related transcription factors are required for coordinated cell cycle progression. Cell cycle (Georgetown, Tex.) 2013, 12, 1762–1772. [Google Scholar] [CrossRef]
- Lei, J.; Hu, D.; Xue, S.; Mao, F.; Obeng, E.; Quan, Y.; Yu, W. HN1L is essential for cell growth and survival during nucleopolyhedrovirus infection in silkworm, Bombyx mori. PLoS One 2019, 14, e0216719. [Google Scholar] [CrossRef]
- Smith, E. R.; Cayrou, C.; Huang, R.; Lane, W. S.; Cote, J.; Lucchesi, J. C. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Molec. Cell. Biol. 2005, 25, 9175–9188. [Google Scholar] [CrossRef]
- Makarov, E.M.; Makarova, O.V.; Achsel, T.; Luhrmann, R. The human homologue of the yeast splicing factor prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein-protein interactions. J. Mol. Biol. 2000, 298, 567–75. [Google Scholar] [CrossRef]
- Han, Y.; Lassen, K.; Monie, D.; Sedaghat, A.R.; Shimoji, S.; Liu, X.; Pierson, T.C.; Margolick, J.B.; Siliciano, R.F.; Siliciano, J.D. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. Journal of virology 2004, 78, 6122–6133. [Google Scholar] [CrossRef]
- Ikeda, T.; Shibata, J.; Yoshimura, K.; Koito, A.; Matsushita, S. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. The Journal of infectious diseases 2007, 195, 716–725. [Google Scholar] [CrossRef]
- Mack, K.D.; Jin, X.; Yu, S.; Wei, R.; Kapp, L.; Green, C.; Herndier, B.; Abbey, N.W.; Elbaggari, A.; Liu, Y.; et al. HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. Journal of acquired immune deficiency syndromes 2003, 33, 308–320. [Google Scholar] [CrossRef]
- Maldarelli, F.; Kearney, A.M.; Palmer, S.; Stephens, R.; Mican, J.; Polis, M.A.; Davey, R.T.; Kovacs, J.; Shao, W.; Rock-Kress, D.; et al. HIV populations are large and accumulate high genetic diversity in a nonlinear fashion. Journal of virology 2013, 87, 10313–10323. [Google Scholar] [CrossRef]
- Flucke, U.; Tops, B.B.; de Saint Aubain Somerhausen, N.; Bras, J.; Creytens, D.H.; Kusters, B.; Groenen, P.J.; Verdijk, M.A.; Suurmeijer, A.J.; Mentzel, T. Presence of C11orf95-MKL2 fusion is a consistent finding in chondroid lipomas: a study of eight cases. Histopathology 2013, 62, 925–930. [Google Scholar] [CrossRef]
- Kobayashi, S.; Taki, T.; Chinen, Y.; Tsutsumi, Y.; Ohshiro, M.; Kobayashi, T.; Matsumoto, Y.; Kuroda, J.; Horiike, S.; Nishida, K.; et al. Identification of IGHCdelta-BACH2 fusion transcripts resulting from cryptic chromosomal rearrangements of 14q32 with 6q15 in aggressive B-cell lymphoma/leukemia. Genes, chromosomes & cancer 2011, 50, 207–216. [Google Scholar]
- Muehlich, S.; Hampl, V.; Khalid, S.; Singer, S.; Frank, N.; Breuhahn, K.; Gudermann, T.; Prywes, R. The transcriptional coactivators megakaryoblastic leukemia 1/2 mediate the effects of loss of the tumor suppressor deleted in liver cancer 1. Oncogene 2012, 31, 3913–3923. [Google Scholar]
- Liu, J.; Sørensen, A.B.; Wang, B.; Wabl, M.; Nielsen, A.L. Pedersen FS. Identification of novel Bach2 transcripts and protein isoforms through tagging analysis of retroviral integrations in B-cell lymphomas. BMC Mol. Biol. 2009, 10, 2199–2210. [Google Scholar] [CrossRef]
- Wang, H.; Bei, L.; Shah, C.A.; Huang, W.; Platanias, L.C.; Eklund, E.A. The E3 ubiquitin ligase Triad1 influences development of Mll-Ell-induced acute myeloid leukemia. Oncogene 2018, 37, 2532–2544. [Google Scholar] [CrossRef]
- Geng, S.; Peng, W.; Wang, X.; Hu, X.; Liang, H.; Hou, J.; Wang, F.; Zhao, G.; Lü, M.; Cui, H. ARIH2 regulates the proliferation, DNA damage and chemosensitivity of gastric cancer cells by reducing the stability of p21 via ubiquitination. Cell Death & Disease, 2022; 13, 1–12. [Google Scholar] [CrossRef]
- Thomason, P.A.; King, J.S.; Insall, R.H. Mroh1, a lysosomal regulator localized by WASH-generated actin. J. Cell. Sci. 2017, 130, 1785–1795. [Google Scholar] [CrossRef]
- Wu, Y.; Gou, Y.; Wang, T.; Li, P.; Li, Y.; Lu, X.; Li, W.; Liu, Z. Exportin XPO6 upregulation activates the TLR2/MyD88/NF-kappaB signaling by facilitating TLR2 mRNA nuclear export in COPD pulmonary monocytes. Int. Immunopharmacol. 2024, 135, 112310. [Google Scholar] [CrossRef]
- Jordan, A.; Defechereux, P.; Verdin, E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 2001, 20, 1726–1738. [Google Scholar] [CrossRef]
- Jordan, A.; Bisgrove, D.; Verdin, E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003, 22, 1868–1877. [Google Scholar] [CrossRef]
- Pearson, R.; Kim, Y.K.; Hokello, J.; Lassen, K.; Friedman, J.; Tyagi, M.; Karn, J. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 2008, 82, 12291–12303. [Google Scholar] [CrossRef]
- Einkauf, K.B.; Lee, G.Q.; Gao, C.; Sharaf, R.; Sun, X.; Hua, S.; Chen, S.M.Y.; Jiang, C.; Lian, X.; Chowdhury, F.Z.; et al. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J. Clin. Invest. 2019, 129, 988–998. [Google Scholar] [CrossRef]
- Jiang, C.; Lian, X.; Gao, C.; Sun, X.; Einkauf, K.B.; Chevalier, J.M.; Chen, S.M.Y.; Hua, S.; Rhee, B.; Chang, K.; et al. Distinct viral reservoirs in individuals with spontaneous control of HIV-1. Nature 2020, 585, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.S.; Ramos, V.; Oliveira, T.Y.; Gaebler, C.; Jankovic, M.; Nussenzweig, M.C.; and Cohn, L.B. Integration features of intact latent HIV-1 in CD4+ T cell clones contribute to viral persistence. J. Exp. Med. 2021, 218, e20211427. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.; Das, B.; Dobrowolski, C.; Karn, J. Multiple histone lysine methyltransferases are required for the establishment and maintenance of HIV-1 latency. MBio 2017, 8, e00133–17. [Google Scholar] [CrossRef]
- Uckelmann, M.; Davidovich, C. Chromatin compaction by Polycomb group proteins revisited. Curr. Opin. Struct. Biol. 2024, 86, 102806. [Google Scholar] [CrossRef]
- Williams, S.A.; Greene, W.C. Regulation of HIV-1 Latency by T-cell Activation. Cytokine 2007, 39, 63–74. [Google Scholar] [CrossRef]
- Bosque, A.; Planelles, V. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 2009, 113, 58–65. [Google Scholar] [CrossRef]
- Tyagi, M.; Pearson, R.J.; Karn, J. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 2010, 84, 6425–6437. [Google Scholar] [CrossRef]
- Han, Y.; Lin, Y.B.; An, W.; Xu, J.; Yang, H.C.; O'Connell, K.; Dordai, D.; Boeke, J.D.; Siliciano, J.D.; Siliciano, R.F. Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 2008, 2, 134–46. [Google Scholar] [CrossRef]
- Shan, L.; Yang, H.C.; Rabi, S.A.; Bravo, H.C.; Shroff, N.S.; Irizarry, R.A.; Zhang, H.; Margolick, J.B.; Siliciano, J.D.; Siliciano, R.F. Influence of host gene transcription level and orientation on HIV-1 latency in a primary-cell model. J. Virol. 2011, 11, 5384–5393. [Google Scholar] [CrossRef]
- Li, G.; Piampongsant, S.; Faria, N.R.; Voet, A.; Pineda-Peña, A.C.; Khouri, R.; Lemey, P.; Vandamme, A.M.; Theys, K. An integrated map of HIV genome-wide variation from a population perspective. Retrovirology 2015, 12, 18. [Google Scholar] [CrossRef]
- Jasinska, A.J. , Apetrei, C., Pandrea, I. Walk on the wild side: SIV infection in African non-human primate hosts-from the field to the laboratory. Front Immunol. 2023, 13, 1060985. [Google Scholar] [CrossRef] [PubMed]
- Casey, L.; Wen, X.; de Noronha, C.M. The functions of the HIV1 protein Vpr and its action through the DCAF1.DDB1.Cullin4 ubiquitin ligase. Cytokine 2010, 51, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, C.C.; Spies, M. Dynamic elements of replication protein A at the crossroads of DNA replication, recombination, and repair. Critical Reviews in Biochemistry and Molecular Biology 2020, 55, 482–507. [Google Scholar] [CrossRef] [PubMed]
- Dueva, R.; Iliakis, G. Replication protein A: a multifunctional protein with roles in DNA replication, repair and beyond. NAR Cancer 2020, 2, zcaa022. [Google Scholar] [CrossRef]
- Sandoval, C.; Nisson, K.; Fregoso, O.I. HIV-1 Vpr induces cell cycle arrest by activating the ATR DNA damage response pathway. PLoS Pathogens 2012, 8, e1002680. [Google Scholar]
- Andersen, J.L.; Planelles, V. The role of Vpr in HIV-1 pathogenesis. Curr. HIV Res. 2005, 3, 43–51. [Google Scholar] [CrossRef]
- Sandoval, C.; Nisson, K.; Fregoso, O.I. HIV-1 Vpr-induced DNA damage activates NF-κB through ATM-NEMO independent of cell cycle arrest. mBio. 2024, 15, e0024024. [Google Scholar] [CrossRef]
- Li, X.; Heyer, W.D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008, 18, 99–113. [Google Scholar] [CrossRef]
- Brandsma, I.; Gent, D.C. Pathway choice in DNA double strand break repair: observations of a balancing act. Genome Integr 2012, 3, 9–18. [Google Scholar] [CrossRef]
- Jäger, S.; Kim, D.Y.; Hultquist, J.F.; Shindo, K.; LaRue, R.S.; Kwon, E.; Li, M.; Anderson, B.D.; Yen, L.; Stanley, D.; et al. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature 2011, 481, 371–5. [Google Scholar] [CrossRef]
- Ramos, F.; Villoria, M.T.; Alonso-Rodriguez, E.; Clemente-Blanco, A. Role of protein phosphatases PP1, PP2A, PP4 and Cdc14 in the DNA damage response. Cell Stress 2019, 3, 70–85. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.; Clemente-Blanco, A. Cell cycle and DNA repair regulation in the damage response: protein phosphatases take over the reins. Int. J. Mol. Sci. 2020, 21, 446–472. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, E.; Bruhn, C.; Peretti, M.; Cassani, C.; Carotenuto, W.V.; Elgendy, M.; Hubassi, G.; Lucca, C.; Bermejo, R.; Varasi, M.; et al. PP2A controls genome integrity by integrating nutrient-sensing and metabolic pathways with the DNA damage response. Mol. Cell. 2017, 67, 266–81. [Google Scholar] [CrossRef]
- Ambjorn, S.M.; Duxin, J.P.; Hertz, E.P.T.; Nasa, I.; Duro, J.; Kruse, T.; Lopez-Mendez, B.; Rymarczyk, B.; Cressey, L.E.; van Overeem Hansen, T.; et al. A complex of BRCA2 and PP2A-B56 is required for DNA repair by homologous recombination. Nat. Commun. 2021, 12, 5748–58. [Google Scholar] [CrossRef] [PubMed]
- Piekna-Przybylska, D.; Sharma, G. .; Maggirwar, S.B.; Bambara, R.A. Deficiency in DNA damage response, a new characteristic of cells infected with latent HIV. Cell Cycle 2017, 16, 968–978. [Google Scholar] [CrossRef]
- Bellon, M.; Nicot, C. Telomere dynamics in immune senescence and exhaustion triggered by chronic viral infection. Viruses 2017, 9, E289. [Google Scholar] [CrossRef]
- Fülöp, T.; Herbein, G.; Cossarizza, A.; Witkowski, J.M.; Frost, E.; Dupuis, G.; et al. Cellular senescence, immunosenescence and HIV. Interdiscip. Top. Gerontol. Geriatr. 2017, 42, 28–46. [Google Scholar]
- Blackburn, E.H. Switching and signaling at the telomere. Cell 2001, 106, 661–73. [Google Scholar]
- Lin, Y.; Damjanovic, A.; Metter, E.J.; Nguyen, H.; Truong, T.; Najarro, K.; Morris, C.; Longo, D.L.; Zhan, M.; Ferrucci, L.; et al. Age-associated telomere attrition of lymphocytes in vivo is co-ordinated with changes in telomerase activity, composition of lymphocyte subsets and health conditions. Clin. Sci. (Lond.) 2015, 128, 367–77. [Google Scholar]
- Rothkamm, K.; Barnard, S.; Moquet, J.; Ellender, M.; Rana, Z.; Burdak-Rothkamm, S. DNA damage foci: meaning and significance. Environ. Mol. Mutagen. 2015, 56, 491–504. [Google Scholar] [CrossRef]
- Alejos, B.; Stella-Ascariz, N.; Montejano, R.; Rodriguez-Centeno, J.; Schwimmer, C.; Bernardino, J.I.; Rodes, B.; Esser, S.; Goujard, C.; Sarmento-Castro, R.; et al. Determinants of blood telomere length in antiretroviral treatment-naive HIV-positive participants enrolled in the NEAT 001/ANRS 143 clinical trial. HIV Med. 2019, 20, 691–8. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, V.C.; Wit, F.W.; Joerink, M.; Maurer, I.; Harskamp, A.M.; Schouten, J.; Prins, M.; van Leeuwen, E.M.; Booiman, T.; Deeks, S.G.; et al. T-cell activation independently associates with immune senescence in HIV infected recipients of long-term antiretroviral treatment. J. Infect. Dis. 2016, 214, 216–225. [Google Scholar] [CrossRef]
- Comandini, A.; Naro, C.; Adamo, R.; Akbar, A.N.; Lanna, A.; Bonmassar, E.; Franzese, O. Molecular mechanisms involved in HIV-1-Tat mediated inhibition of telomerase activity in human CD4(+) T lymphocytes. Mol. Immunol. 2013, 54, 181–92. [Google Scholar] [CrossRef]
- Stella-Ascariz, N.; Montejano, R.; Rodriguez-Centeno, J.; Alejos, B.; Schwimmer, C.; Bernardino, J.I.; Rodes, B.; Allavena, C.; Hoffmann, C.; Gisslén, M.; et al. Blood telomere length changes after ritonavir-boosted darunavir combined with raltegravir or tenofovir-emtricitabine in antiretroviral-naïve adults infected with HIV-1. J. Infect. Dis. 2018, 218, 1523–30. [Google Scholar] [CrossRef]
- Raffenberg, M.; Engel, T.; Schoepf, I.C.; Kootstra, N.A.; Reiss, P.; Braun, D.L.; Thorball, C.W.; Fellay, J.; Kouyos, R.D.; Ledergerber, B.; et al. Impact of delaying antiretroviral treatment during primary human immunodeficiency virus infection on telomere length. J. Infect. Dis. 2021, 224, 1775–84. [Google Scholar] [CrossRef]
- Montejano, R.; Stella-Ascariz, N.; Monge, S.; Bernardino, J.I.; Pérez-Valero, I.; Montes, M.L.; Valencia, E.; Martín-Carbonero, L.; Moreno, V.; González-Garcia, J.; et al. Impact of nucleos(t)ide reverse transcriptase inhibitors on blood telomere length changes in a prospective cohort of aviremic HIV-infected adults. J. Infect. Dis. 2018, 218, 1531–40. [Google Scholar] [CrossRef]
- Kaushal, S.; Landay, A.L.; Lederman, M.M.; Connick, E.; Spritzler, J.; Kuritzkes, D.R.; Kessler, H.; Levine, B.L.; St Louis, D.C.; June, C. H. Increases in T cell telomere length in HIV infection after antiretroviral combination therapy for HIV-1 infection implicate distinct population dynamics in CD4+ and CD8+ T cells. Clin. Immunol. 1999, 92, 14–24. [Google Scholar] [CrossRef]
- Hukezalie, K.R.; Thumati, N.R.; Côté, H.C.; Wong, J.M. In vitro and ex vivo inhibition of human telomerase by anti-HIV nucleoside reverse transcriptase inhibitors (NRTIs) but not by non-NRTIs. PLoS One 2012, 7. [Google Scholar] [CrossRef]
- Stella-Ascariz, N.; Montejano, R.; Pintado-Berninches, L.; Monge, S.; Bernardino, J.I.; Pérez-Valero, I.; Montes, M.L.; Mingorance, J.; Perona, R.; Arribas, J.R. Differential effects of tenofovir, abacavir, emtricitabine, and darunavir on telomerase activity in vitro. J. Acquir. Immune Defic. Syndr. 2017, 74, 91–4. [Google Scholar] [CrossRef]
- Leeansyah, E.; Cameron, P.U.; Solomon, A.; Tennakoon, S.; Velayudham, P.; Gouillou, M.; Spelman, T.; Hearps, A.; Fairley, C.; Smit, deV. ; et al. Inhibition of telomerase activity by human immunodeficiency virus (HIV) nucleos(t)ide reverse transcriptase inhibitors: a potential factor contributing to HIV-associated accelerated aging. J. Infect. Dis. 2013, 207, 1157–65. [Google Scholar] [CrossRef]
- Lombardi, F.; Sanfilippo, A.; Fabbiani, M.; Borghetti, A.; Ciccullo, A.; Tamburrini, E.; Di Giambenedetto, S. Blood telomere length gain in people living with HIV switching to dolutegravir plus lamivudine versus continuing triple regimen: a longitudinal, prospective, matched, controlled study. J. Antimicrob. Chemother. 2023, 9, 2315–2322. [Google Scholar] [CrossRef] [PubMed]
- Dalzini, A.; Ballin, G.; Dominguez-Rodriguez, S.; Rojo, P.; Petrara, M. R.; Foster, C.; Cotugno, N.; Ruggiero, A.; Nastouli, E.; Klein, N.; et al. Size of HIV-1 reservoir is associated with telomere shortening and immunosenescence in early-treated European children with perinatally acquired HIV-1. J. Int. AIDS Soc. 2021, 11, e25847. [Google Scholar] [CrossRef] [PubMed]
- Appay, V.; Sauce, D. Assessing immune aging in HIV-infected patients. Virulence 2017, 8, 529–38. [Google Scholar] [CrossRef]
- Jose, S.S.; Bendickova, K.; Kepak, T.; Krenova, Z.; Fric, J. Chronic inflammation in immune aging: role of pattern recognition receptor crosstalk with the telomere complex? Front. Immunol. 2017, 8, 1078. [Google Scholar] [CrossRef]
- Rodier, F.; Coppé, J.P.; Patil, C.K.; Hoeijmakers, W.A.; Muñoz, D.P.; Raza, S.R.; Freund, A.; Campeau, E.; Davalos, A.R.; Campisi, J. Persistent DNA damage signaling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 2009, 11, 973–9. [Google Scholar] [CrossRef]
- Van Epps, P.; Kalayjian, R.C. Human immunodeficiency virus and aging in the era of effective antiretroviral therapy. Infect. Dis. Clin. North Am. 2017, 31, 791–810. [Google Scholar] [CrossRef]
- Blanco, J.R.; Jarrin, I.; Martinez, A.; Siles, E.; Larrayoz, I.M.; Cañuelo, A.; Gutierrez, F.; Gonzalez-Garcia, J.; Vidal, F.; Moreno, S.; et al. Shorter telomere length predicts poorer immunological recovery in virologically suppressed HIV-1-infected patients treated with combined antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 2015, 68, 21–9. [Google Scholar] [CrossRef]
- Shankarappa, R.; Gupta, P.; Learn Jr, G.H.; Rodrigo, A.G.; Rinaldo Jr, C.R.; Gorry, M.C.; Mullins, J.I.; Nara, P.L.; Ehrlich, G.D. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology 1998, 241, 251–259. [Google Scholar] [CrossRef]
- Bailey, J.R.; Sedaghat, A.R.; Kieffer, T.; Brennan, T.; Lee, P.K.; Wind-Rotolo, M.; Haggerty, C.M.; Kamireddi, A.R.; Liu, Y.; Lee, J.; et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. Journal of virology 2006, 80, 6441–6457. [Google Scholar] [CrossRef]
- Kearney, M.F.; Spindler, J.; Shao, W.; Yu, S.; Anderson, E.M.; O'Shea, A.; Rehm, C.; Poethke, C.; Kovacs, N.; Mellors, J.W.; et al. Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy. PLoS pathogens 2014, 10, e1004010. [Google Scholar] [CrossRef]
- Besson, G.J.; Lalama, C.M.; Bosch, R.J.; Gandhi, R.T.; Bedison, M.A.; Aga, E.; Riddler, S.A.; McMahon, D.K.; Hong, F.; Mellors, J.W. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin. Infect. Dis. 2014, 59, 1312–1321. [Google Scholar] [CrossRef] [PubMed]
- Pollack, R.A.; Jones, R.B.; Pertea, M.; Bruner, K.M.; Martin, A.R.; Thomas, A.S.; Capoferri, A.A.; Beg, S.A.; Huang, S.H.; Karandish, S.; et al. Defective HIV-1 Proviruses Are Expressed and Can Be Recognized by Cytotoxic T Lymphocytes, which Shape the Proviral Landscape. Cell Host Microbe, 2017, 21, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.C.; Shan, L.; Hosmane, N.N.; Wang, J.; Laskey, S.B.; Rosenbloom, D.I.; Lai, J.; Blankson, J.N.; Siliciano, J.D.; Siliciano, R.F. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013, 155, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Musick, A.; Spindler, J.; Boritz, E.; Perez, L.; Crespo-Velez, D.; Patro, S.C.; Sobolewski, M.D.; Bale, M.J.; Reid, C.; Keele, B.F.; et al. HIV Infected T Cells Can Proliferate in vivo Without Inducing Expression of the Integrated Provirus. Front. Microbiol. 2019, 10, 2204. [Google Scholar] [CrossRef]
- Coffin, J.M.; Bale, M.J.; Wells, D.; Guo, S.; Luke, B.; Zerbato, J.M.; Sobolewski, M.D.; Sia, T.; Shao, W.; Wu, X.; et al. Integration in oncogenes plays only a minor role in determining the in vivo distribution of HIV integration sites before or during suppressive antiretroviral therapy. PLoS Pathog. 2021, 17, e1009141. [Google Scholar] [CrossRef]
- Einkauf, K.B.; Osborn, M.R.; Gao, C.; Sun, W.; Sun, X.; Lian, X.; Parsons, E.M.; Gladkov, G.T.; Seiger, K.W.; Blackmer, J.E.; et al. Parallel analysis of transcription, integration, and sequence of single HIV-1 proviruses. Cell 2022, 185, 266–282.e15. [Google Scholar] [CrossRef]
- Cole, B.; Lambrechts, L.; Gantner, P.; Noppe, Y.; Bonine, N.; Witkowski, W.; Chen, L.; Palmer, S.; Mullins, J.I.; Chomont, N.; et al. In-depth single-cell analysis of translation-competent HIV-1 reservoirs identifies cellular sources of plasma viremia. Nat. Commun. 2021, 12, 3727. [Google Scholar] [CrossRef]
- Lian, X.; Seiger, K.W.; Parsons, E.M.; Gao, C.; Sun, W.; Gladkov, G.T.; Roseto, I.C.; Einkauf, K.B.; Osborn, M.R.; Chevalier, J.M.; et al. Progressive transformation of the HIV-1 reservoir cell profile over two decades of antiviral therapy. Cell Host Microbe 2023, 31, 83–96.e5. [Google Scholar] [CrossRef]
- Matsui, T.; Leung, D.; Miyashita, H.; Maksakova, I.A.; Miyachi, H.; Kimura, H.; Tachibana, M.; Lorincz, M.C.; Shinkai, Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 2010, 464, 927–931. [Google Scholar] [CrossRef]
- Schultz, D.C.; Ayyanathan, K.; Negorev, D.; Maul, G.G.; Rauscher, F.J. , 3rd. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002, 16, 919–932. [Google Scholar] [CrossRef]
- Halvas, E.K.; Joseph, K.W.; Brandt, L.D.; Guo, S.; Sobolewski, M.D.; Jacobs, J.L.; Tumiotto, C.; Bui, J.K.; Cyktor, J.C.; Keele, B.F.; et al. HIV-1 viremia not suppressible by antiretroviral therapy can originate from large T cell clones producing infectious virus. J. Clin. Invest. 2020, 11, 5847–5857. [Google Scholar] [CrossRef] [PubMed]
- Marini, B.; Kertesz-Farkas, A.; Ali, H.; Lucic, B.; Lisek, K.; Manganaro, L.; Pongor, S.; Luzzati, R.; Recchia, A.; Mavilio, F.; et al. Nuclear architecture dictates HIV-1 integration site selection. Nature, 2015; 521, 227–231. [Google Scholar] [CrossRef]
- Matreyek, K.A.; Yücel, S.S.; Li, X.; Engelman, A. Nucleoporin Nup153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog 2013, 9, e1003693. [Google Scholar] [CrossRef]
- Ocwieja, K.E.; Brady, T.L.; Ronen, K.; Huegel, A.; Roth, S.L.; Schaller, T.; James, L.C.; Towers, G.J.; Young, J.A.; Chanda, S.K.; et al. HIV integration targeting: a pathway involving transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog 2011, 7, e1001313. [Google Scholar] [CrossRef]
- Koh, Y.; Wu, X.; Ferris, A.L.; Matreyek, K.A.; Smith, S.J.; Lee, K.; KewalRamani, V.N.; Hughes, S.H.; Engelman, A. Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. J. Virol. 2013, 87, 648–658. [Google Scholar] [CrossRef]
- Di Nunzio, F.; Fricke, T.; Miccio, A.; Valle-Casuso, J.C.; Perez, P.; Souque, P.; Rizzi, E.; Severgnini, M.; Mavilio, F.; Charneau, P.; et al. Nup153 and Nup98 bind the HIV-1 core and contribute to the early steps of HIV-1 replication. Virology 2013, 440, 8–18. [Google Scholar] [CrossRef]
- Achuthan, V.; Perreira, J.M.; Sowd, G.A.; Puray-Chavez, M.; McDougall, W.M.; Paulucci-Holthauzen, A.; Wu, X.; Fadel, H.J.; Poeschla, E.M.; Multani, A.S.; et al. Capsid-CPSF6 interaction licenses nuclear HIV-1 trafficking to sites of viral DNA integration. Cell Host Microbe 2018, 24, 392–404. [Google Scholar] [CrossRef]
- Briand, N.; Collas, P. Lamina-associated domains: peripheral matters and internal affairs. Genome Biol. 2020, 1, 85. [Google Scholar] [CrossRef]
- Alagna, N.S.; Thomas, T.I.; Wilson, K.L.; Reddy, K.L. Choreography of lamina-associated domains: structure meets dynamics. FEBS Lett. 2023 Nov;597(22):2806-2822. [CrossRef]
- Bester, S.M.; Wei, G.; Zhao, H.; Adu-Ampratwum, D.; Iqbal, N.; Courouble, V.V.; Francis, A.C.; Annamalai, A.S.; Singh, P.K.; Shkriabai, N.; et al. Structural and mechanistic bases for a potent HIV-1 capsid inhibitor. Science 2020, 6514, 360–364. [Google Scholar] [CrossRef]
- Emerson, R.O.; Thomas, J.H. Gypsy and the birth of the SCAN domain. J. Virol. 2011, 22, 12043–52. [Google Scholar] [CrossRef]
- Williams, A.J.; Blacklow, S.C.; Collins, T. The zinc finger-associated SCAN box is a conserved oligomerization domain. Mol. Cell. Biol. 1999, 19, 8526–8535. [Google Scholar] [CrossRef]
- Bellefroid, E.J.; Poncelet, D.A.; Lecocq, P.J.; Revelant, O.; Martial, J.A. The evolutionarily conserved Krüppel-associated box domain defined a subfamily of eu-karyotic multifingered proteins. Proc. Natl. Acad. Sci. U. S. A. 1991, 88, 3608–3612. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.; Skorupka, K.A.; Pak, A.J.; Ganser-Pornillos, B.K.; Pornillos, O.; Voth, G.A. TRIM5α self-assembly and compartmentalization of the HIV-1 viral capsid. Nat. Commun. 2020, 11, 1307–1316. [Google Scholar] [CrossRef]
- Jiang, C.; Lian, X.; Gao, C.; Sun, X.; Einkauf, K.B.; Chevalier, J.M.; Chen, S.M.Y.; Hua, S.; Rhee, B.; Chang, K.; et al. Distinct viral reservoirs in individuals with spontaneous control of HIV-1. Nature 2020, 585, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.; Jolicoeur, P. Retroviral pathogenesis. in Retrovirus, JM Coffin, SH Hughes, HE Varmus, Eds. (Cold Spring Harbor Laboratory Press, 1997), pp. 475–585.
- Neil, J.C.; Cameron, E.R. Retroviral insertion sites and cancer: fountain of all knowledge? Cancer Cell 2002, 4, 253–5. [Google Scholar] [CrossRef] [PubMed]
- McNally, G.A. HIV and Cancer: An Overview of AIDS-Defining and Non-AIDS-Defining Cancers in Patients With HIV. Clin. J. Oncol. Nurs. 2019, 3, 327–331. [Google Scholar] [CrossRef]
- Biggar, R.J.; Engels, E.A.; Frisch, M.; Goedert, J.J.; AIDS Cancer Match Registry Study Group. Risk of T-cell lymphomas in persons with AIDS. J. Acquir. Immune Defic. Syndr. 2001, 4, 371–6. [Google Scholar] [CrossRef]
- Herndier, B.G.; Shiramizu, B.T.; Jewett, N.E.; Aldape, K.D.; Reyes, G.R.; McGrath, M.S. ; Acquired immunodeficiency syndrome-associated T-cell lymphoma: Evidence for human immunodeficiency virus type 1-associated T-cell transformation. Blood 1992, 79, 1768–1774. [Google Scholar]
- Shiramizu, B.; Herndier, B.G.; McGrath, M.S. ; Identification of a common clonal human immunodeficiency virus integration site in human immunodeficiency virus-associated lymphomas. Cancer Res. 1994, 54, 2069–2072. [Google Scholar]
- Katano, H.; Sato, Y.; Hoshino, S.; Tachikawa, N.; Oka, S.; Morishita, Y.; Ishida, T.; Watanabe, T.; Rom, W.N.; Mori, S.; et al. Integration of HIV-1 caused STAT3-associated B cell lymphoma in an AIDS patient. Microbes Infect. 2007, 9, 1581–1589. [Google Scholar] [CrossRef]
- Tolomeo, M.; Cascio, A. The Multifaced Role of STAT3 in Cancer and Its Implication for Anticancer Therapy. Int. J. Mol. Sci. 2021, 2, 603. [Google Scholar] [CrossRef]
- Mellors, J.W.; Guo, S.; Naqvi, A.; Brandt, L.D.; Su, L.; Sun, Z.; Joseph, K.W.; Demirov, D.; Halvas, E.K.; Butcher, D.; Scott, B.; et al. Insertional activation of STAT3 and LCK by HIV-1 proviruses in T cell lymphomas. Sci. Adv. 2021, 42, eabi8795. [Google Scholar] [CrossRef]
- Almodovar, S.; Knight, R.; Allshouse, A.A.; Roemer, S.; Lozupone, C.; McDonald, D.; Widmann, J.; Voelkel, N.F.; Shelton, R.J.; Suarez, E.B.; et al. Human Immunodeficiency Virus nef signature sequences are associated with pulmonary hypertension. AIDS Res. Hum. Retrovir. 2012, 28, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Musinova, Y.R.; Sheval, E.V.; Dib, C.; Germini, D.; Vassetzky, Y.S. Functional roles of HIV-1 Tat protein in the nucleus. Cell. Mol. Life Sci. 2016, 3, 589–601. [Google Scholar] [CrossRef]
- Manes, T.L.; Simenauer, A.; Geohring, J.L.; Flemming, J.; Brehm, M.; Cota-Gomez, A. The HIV-Tat protein interacts with Sp3 transcription factor and inhibits its binding to a distal site of the sod2 promoter in human pulmonary artery endothelial cells. Free Radic. Biol. Med. 2020, 147, 102–113. [Google Scholar] [CrossRef]
- Col, E.; Caron, C.; Chable-Bessia, C.; Legube, G.; Gazzeri, S.; Komatsu, Y.; Yoshida, M.; Benkirane, M.; Trouche, D.; Khochbin, S. HIV-1 Tat targets Tip60 to impair the apoptotic cell response to genotoxic stresses. EMBO J. 2005, 24, 2634–2645. [Google Scholar] [CrossRef]
- Zhang, S.M.; Song, M.; Yang, T.Y.; Fan, R.; Liu, X.D.; Zhou, P.K. HIV-1 tat impairs cell cycle control by targeting the Tip60, Plk1 and cyclin B1 ternary complex. Cell Cycle 2012, 11, 1217–1234. [Google Scholar] [CrossRef]
- Martina, C.; Fabienne, H.; Vesco, M.; Edwige, C.; Cécile, C.; Stefan, D.; Saadi, K. Control of the Histone-Acetyltransferase Activity of Tip60 by the HIV-1 Transactivator Protein, Tat. Biochemistry 1999, 38, 8826–8830. [Google Scholar]
- Sun, Y.; Jiang, X.; Chen, S.; Fernandes, N.; Price, B.D. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. USA 2005, 102, 13182–13187. [Google Scholar] [CrossRef]
- Baur, A. Functions of the HIV-1 Nef protein. Curr. Drug Targets Immune Endocr. Metabol. Disord. 2004, 4, 309–13. [Google Scholar] [CrossRef]
- Wang, T.; Green, L.A.; Gupta, S.K.; Kim, C.; Wang, L.; Almodovar, S.; Flores, S.C.; Prudovsky, I.A.; Jolicoeur, P.; Liu, Z.; et al. Transfer of intracellular HIV Nef to endothelium causes endothelial dysfunction. PLoS One. 2014, 3, e91063. [Google Scholar] [CrossRef]
- Wolf, D.; Witte, V.; Laffert, B.; Blume, K.; Stromer, E.; Trapp, S.; d'Aloja, P.; Schürmann, A.; Baur, A.S. HIV-1 Nef associated PAK and PI3-kinases stimulate Akt-independent Bad-phosphorylation to induce anti-apoptotic signals. Nat. Med. 2001, 11, 1217–24. [Google Scholar] [CrossRef] [PubMed]
- Rednor, S.J.; Ross, M.J. Molecular Mechanisms of Injury in HIV-Associated Nephropathy. Front. Med. (Lausanne). 2018, 7, 5–177. [Google Scholar] [CrossRef] [PubMed]
- Bruggeman, L.A.; Ross, M.D.; Tanji, N.; Cara, A.; Dikman, S.; Gordon, R.E.; Burns, G.C.; D'Agati, V.D.; Winston, J.A.; Klotman, M.E.; et al. Renal epithelium is a previously unrecognized site of HIV-1 infection. J. Am. Soc. Nephrol. 2000, 11, 2079–87. [Google Scholar] [CrossRef]
- Eitner, F.; Cui, Y.; Hudkins, K.L.; Stokes, M.B.; Segerer, S.; Mack, M.; Lewis, P.L.; Abraham, A.A.; Schlöndorff, D.; Gallo, G.; et al. Chemokine receptor CCR5 and CXCR4 expression in HIV-associated kidney disease. J. Am. Soc. Nephrol. 2000, 11, 856–867. [Google Scholar] [CrossRef]
- Ray, P.E.; Liu, X.H.; Henry, D.; Dye 3rd, L.; Xu, L.; Orenstein, J.M.; Schuztbank, T.E. Infection of human primary renal epithelial cells with HIV-1 from children with HIV-associated nephropathy. Kidney Int. 1998, 53, 1217–29. [Google Scholar]
- Chen, P.; Chen, B.K.; Mosoian, A.; Hays, T.; Ross, M.J.; Klotman, P.E.; Klotman, M.E. Virological synapses allow HIV-1 uptake and gene expression in renal tubular epithelial cells. J. Am. Soc. Nephrol. 2011, 22, 496–507. [Google Scholar] [CrossRef]
- Li, J.; Das, J.R.; Tang, P.; Han, Z.; Jaiswal, J.K.; Ray, P.E. Transmembrane TNF- facilitates HIV-1 infection of podocytes cultured from children with HIV-associated nephropathy. J. Am. Soc. Nephrol. 2016, 28, 862–75. [Google Scholar] [CrossRef]
- Ikura, T.; Tashiro, S.; Kakino, A.; Shima, H.; Jacob, N.; Amunugama, R.; Yoder, K.; Izumi, S.; Kuraoka, I.; Tanaka, K.; et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 2007, 20, 7028–40. [Google Scholar] [CrossRef]
- Prabhu, K.S.; Kuttikrishnan, S.; Ahmad, N.; Habeeba, U.; Mariyam, Z.; Suleman, M.; Bhat, A.A.; Uddin, S. H2AX: A key player in DNA damage response and a promising target for cancer therapy. Biomed. Pharmacother. 2024, 175, 116663. [Google Scholar] [CrossRef]
- Rosenstiel, P.E.; Chan, J.; Snyder, A.; Planelles, V.; D'Agati, V.D.; Klotman, P.E.; Klotman, M.E. HIV-1 Vpr activates the DNA damage response in renal tubule epithelial cells. AIDS 2009, 15, 2054–6. [Google Scholar] [CrossRef]
- Spudich, S.; González-Scarano, F. HIV-1-related central nervous system disease: current issues in pathogenesis, diagnosis, and treatment. Cold Spring Harb. Perspect. Med. 2012, 2, a007120. [Google Scholar] [CrossRef] [PubMed]
- Boisse, L.; Gill, M.J.; Power, C. HIV infection of the central nervous system: clinical features and neuropathogenesis. Neurol. Clin. 2008, 26, 799–819. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Power, C. Regulation of neural cell survival by HIV-1 infection. Neurobiol. Dis. 2012, 21, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Chopp, M.; Chan, P.H.; Hsu, C.Y.; Cheung, M.E.; Jacobs, T.P. DNA damage and repair in central nervous system injury: National Institute of Neurological Disorders and Stroke Workshop Summary. Stroke. 1996, 27, 363–9. [Google Scholar] [CrossRef]
- Madabhushi, R.; Pan, L.; Tsai, L.H. DNA damage and its links to neurodegeneration. Neuron 2014, 83, 266–282. [Google Scholar] [CrossRef]
- Madabhushi, R.; Gao, F.; Pfenning, A.R.; Pan, L.; Yamakawa, S.; Seo, J.; Rueda, R.; Phan, T.X.; Yamakawa, H.; Pao, P.C.; et al. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 2015, 161, 1592–605. [Google Scholar] [CrossRef]
- James, T.; Nonnemacher, M.R.; Wigdahl, B.; Krebs, F.C. Defining the roles for Vpr in HIV-1-associated neuropathogenesis. J. Neurovirol. 2016, 22, 403–15. [Google Scholar] [CrossRef]
- Crewe, M.; Madabhushi, R. Topoisomerase-mediated DNA damage in neurological disorders. Front Aging Neurosci. 2021, 13, 751742–59. [Google Scholar] [CrossRef]
- Madabhushi, R. The Roles of DNA Topoisomerase IIβ in Transcription. Int. J. Mol. Sci. 2018, 19, 1917. [Google Scholar] [CrossRef]
- McKinnon, P.J. DNA repair deficiency and neurological disease. Nat. Rev. Neurosci. 2009, 10, 100–112. [Google Scholar] [CrossRef]
- McKinnon, P.J. Maintaining genome stability in the nervous system. Nat. Neurosci. 2013, 16, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Shull, E.R.; Lee, Y.; Nakane, H.; Stracker, T.H.; Zhao, J.; Russell, H.R.; Petrini, J.H.; McKinnon, P.J. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes Dev. 2009, 23, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Hollinger, A.; Cueni, N.; Marzolini, C.; Dickenmann, M.; Landmann, E.; Battegay, M.; Martinez, A. E,. Siegemund, M.; Leuppi-Taegtmeyer, A. Lactic acidosis and hyperlactatemia associated with lamivudine accumulation and sepsis in a kidney transplant recipient-a case report and review of the literature. AIDS Res. Ther. 2021, 1, 56. [Google Scholar]
- Oliveira, N.M.; Ferreira, F.A.; Yonamine, R.Y.; Chehter, E.Z. Antiretroviral drugs and acute pancreatitis in HIV/AIDS patients: is there any association? A literature review. Einstein (Sao Paulo). 2014, 1, 112–119. [Google Scholar] [CrossRef]
- Jao, J.; Wyatt, C.M. Antiretroviral medications: adverse effects on the kidney. Adv. Chronic Kidney Dis. 2010, 1, 72–82. [Google Scholar] [CrossRef]
- Hitchcock, A.M.; Kufel, W.D.; Dwyer, K.A.M.; Sidman, E.F. Lenacapavir: A novel injectable HIV-1 capsid inhibitor. Int. J. Antimicrob. Agents 2024, 1, 107009. [Google Scholar] [CrossRef]
- Engelman, A.N. HIV Capsid and Integration Targeting. Viruses 2021, 1, 125. [Google Scholar] [CrossRef]
- Mounzer, K.; Slim, J.; Ramgopal, M.; Hedgcock, M.; Bloch, M.; Santana, J.; Mendes, I.; Zhang, X.; Sklar, P.; Montezuma-Rusca, J.M.; et al. Efficacy and safety of bictegravir plus lenacapavir: 48-week outcomes in virologically suppressed people with HIV-1 on complex antiretroviral regimens at baseline. AIDS 2024, , Munich. Oral abstract OAB2602. 22–26 July.
- Mounzer, K. , Slim, J.; Ramgopal, M.; Hedgcock, M.; Bloch, M.; Santana, J.; Mendes, I.; Guo, Y.; Arora, P.; Montezuma-Rusca, J.M. et al. Phase 2 study of switch to daily BIC + LEN in individuals on a complex HIV treatment regimen. CROI 2024, Denver.
- Mounzer, K. , Slim, J.; Ramgopal, M., Hedgcock, M., Bloch, M., Santana, J., Mendes, I., Guo, Y., Arora, P., Eds.; Montezuma-Rusca, J.M. et al. Efficacy and Safety of Switching to Daily Bictegravir Plus Lenacapavir From a Complex Human Immunodeficiency Virus Treatment Regimen: A Randomized, Open-Label, Multicenter Phase 2 Study (ARTISTRY-1). Clin. Infect. Dis. 2024. [Google Scholar]
- Doan, J.; Brunzo-Hager, S.; Satterly, B.; Cory, T.J. Expanding therapeutic options: lenacapavir + bictegravir as a potential treatment for HIV. Expert Opin. Pharmacother. 2023, 18, 1949–1956. [Google Scholar] [CrossRef]
- Pak, A.J.; Grime, J.M.A.; Yu, A.; Voth, GA. Off-pathway assembly: a broad-spectrum mechanism of action for drugs that undermine controlled HIV-1 viral capsid formation. J. Am. Chem. Soc. 2019, 141, 10214–10224. [Google Scholar] [CrossRef]
- Gruell, H.; Gunst, J.D.; Cohen, Y.Z.; Pahus, M.H.; Malin, J.J.; Platten, M.; Millard, K.G.; Tolstrup, M.; Jones, R.B.; Conce Alberto, W.D.; et al. Effect of 3BNC117 and romidepsin on the HIV-1 reservoir in people taking suppressive antiretroviral therapy (ROADMAP): a randomised, open-label, phase 2A trial. Lancet Microbe 2022, 3, e203–e214. [Google Scholar] [CrossRef]
- Maxwell, J.W.; Falcinelli, S.D.; Nefedov, A.; Dorfmeier, C.; Wu, G.; Dewey, M.; Webber, A.L.; Archin, N.M.; Margolis, D.M.; Hazuda, D.J.; et al. Cellular Gene Modulation of HIV-Infected CD4 T Cells in Response to Serial Treatment with the Histone Deacetylase Inhibitor Vorinostat. J. Virol. 2020, 13, e00351–20. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, A.P.; Badley, A.D. Prime, shock and kill: BCL-2 inhibition for HIV cure. Front. Immunol. 2022, 13, 1033609. [Google Scholar] [CrossRef] [PubMed]
- Ahlenstiel, C.L.; Symonds, G.; Kent, S.J.; Kelleher, A.D. Block and Lock HIV Cure Strategies to Control the Latent Reservoir. Front. Cell. Infect. Microbiol. 2020, 10, 424. [Google Scholar] [CrossRef]
- Mousseau, G.; Kessing, C.F.; Fromentin, R.; Trautmann, L.; Chomont, N.; Valente, S.T. The Tat Inhibitor Didehydro-Cortistatin A Prevents HIV-1 Reactivation From Latency. mBio, 2015, 4, e00465. [Google Scholar] [CrossRef]
- Li, C.; Mousseau, G.; Valente, S.T. Tat Inhibition by Didehydro-Cortistatin A Promotes Heterochromatin Formation at the HIV-1 Long Terminal Repeat. Epigenet. Chromatin 2019, 1, 23. [Google Scholar] [CrossRef]
- Kessing, C.F.; Nixon, C.C.; Li, C.; Tsai, P.; Takata, H.; Mousseau, G.; Ho, P.T.; Honeycutt, J.B.; Fallahi, M.; Trautmann, L.; et al. In Vivo Suppression of HIV Rebound by Didehydro-Cortistatin A, a "Block-And-Lock" Strategy for HIV-1 Treatment. Cell Rep. 2017, 3, 600–11. [Google Scholar] [CrossRef]
- Bhowmik, R.; Chaubey, B. CRISPR/Cas9: a tool to eradicate HIV-1. AIDS Res. Ther. 2022, 1, 58. [Google Scholar] [CrossRef]
- Dash, P.K.; Chen, C.; Kaminski, R.; Su, H.; Mancuso, P.; Sillman, B.; Zhang, C.; Liao, S.; Sravanam, S.; Liu, H.; et al. CRISPR editing of CCR5 and HIV-1 facilitates viral elimination in antiretroviral drug-suppressed virus-infected humanized mice. Proc. Natl. Acad. Sci. U. S. A. 2023, 19, e2217887120. [Google Scholar] [CrossRef]
- Herskovitz, J.; Hasan, M.; Patel, M.; Blomberg, W.R.; Cohen, J.D.; Machhi, J.; Shahjin, F.; Mosley, R.L.; McMillan, J.; Kevadiya, B.D.; et al. CRISPR-Cas9 Mediated Exonic Disruption for HIV-1 Elimination. EBioMedicine 2021, 73, 103678. [Google Scholar] [CrossRef]
- Cohrt, K.O., News. Excision’s EBT-101 demonstrates safety in clinical trial but does not cure HIV - The 2nd CRISPR Medicine Conference 2025, -11, Copenhagen, Denmark. 7 April.
- Gupta, R.K.; Peppa, D.; Hill, A.L.; Gálvez, C.; Salgado, M.; Pace, M.; McCoy, L.E.; Griffith, S.A.; Thornhill, J.; Alrubayyi, A. Evidence for HIV-1 cure after CCR5Δ32/Δ32 allogeneic haemopoietic stem-cell transplantation 30 months post analytical treatment interruption: a case report. The Lancet HIV 2020, 7, 5–e340. [Google Scholar] [CrossRef]
- Hütter, G.; Nowak, D.; Mossner, M.; Ganepola, S.; Müßig, A.; Allers, K.; Schneider, T.; Homann, J.; Kücherer, C.; Blau, O. "Long-term remission and HIV cure in a patient with a CCR5-delta32/Delta32 mutation after stem cell transplantation. " New England Journal of Medicine 2009, 360, 692–698. [Google Scholar] [PubMed]
- Li, J.Z.; Blankson, J.N. How elite controllers and posttreatment controllers inform our search for an HIV-1 cure. J. Clin. Invest. 2021, 11, e149414. [Google Scholar]
- Derbalah, A.; Karpick, H.C.; Maize, H.; Skersick, P.; Cottrell, M.; Rao, G.G. Role of islatravir in HIV treatment and prevention: an update. Curr. Opin. HIV AIDS 2022, 4, 240–246. [Google Scholar] [CrossRef]
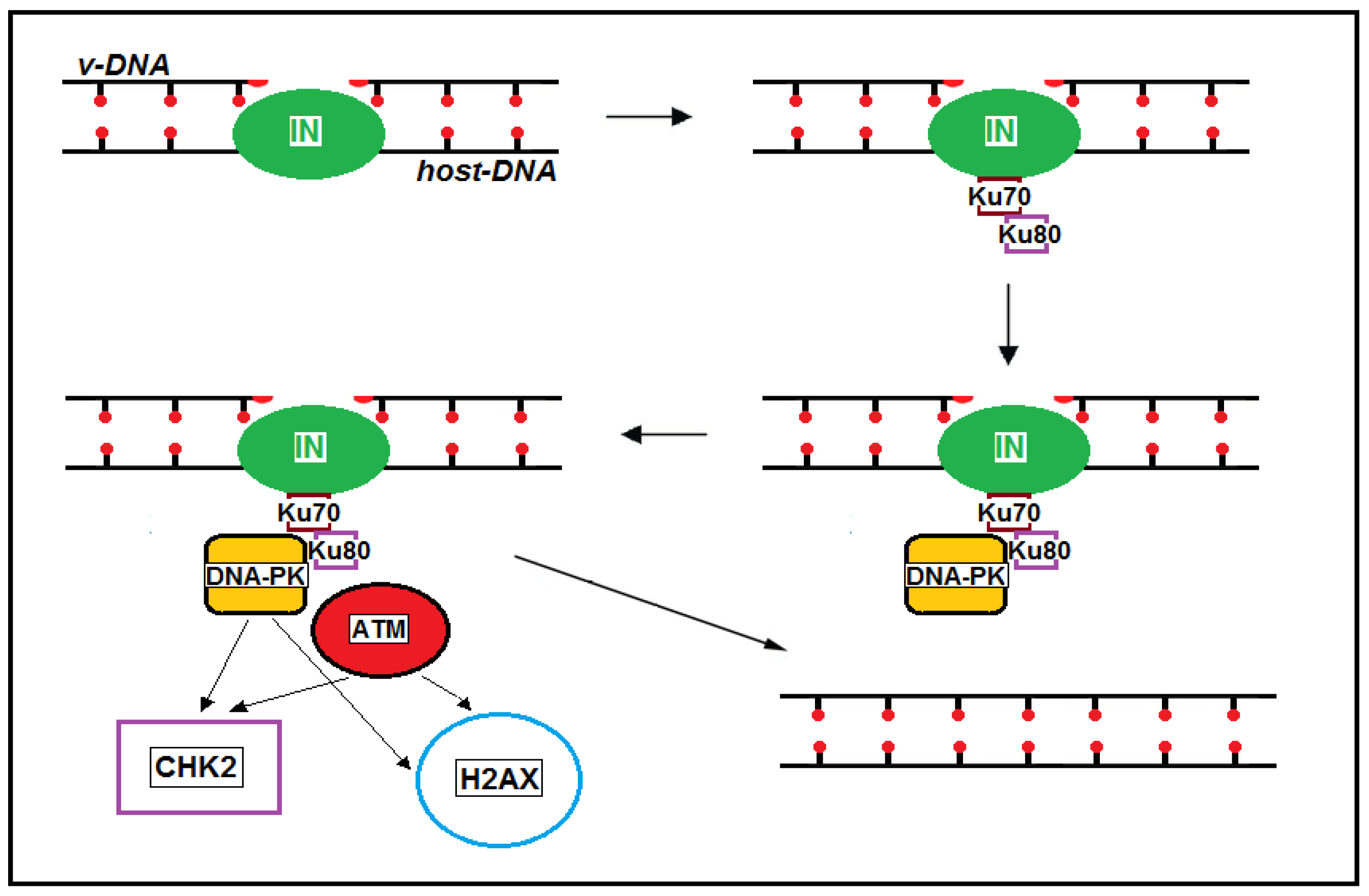
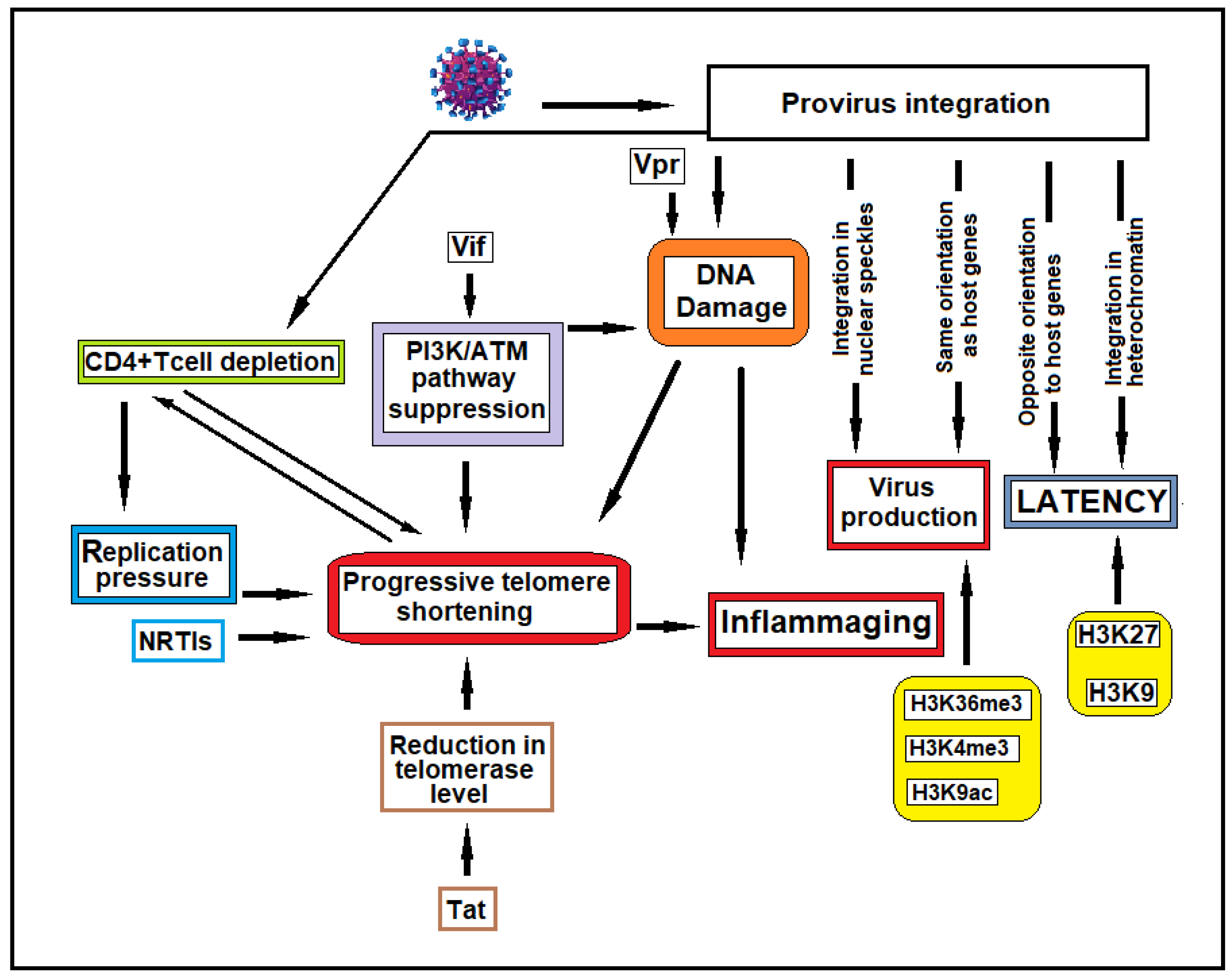
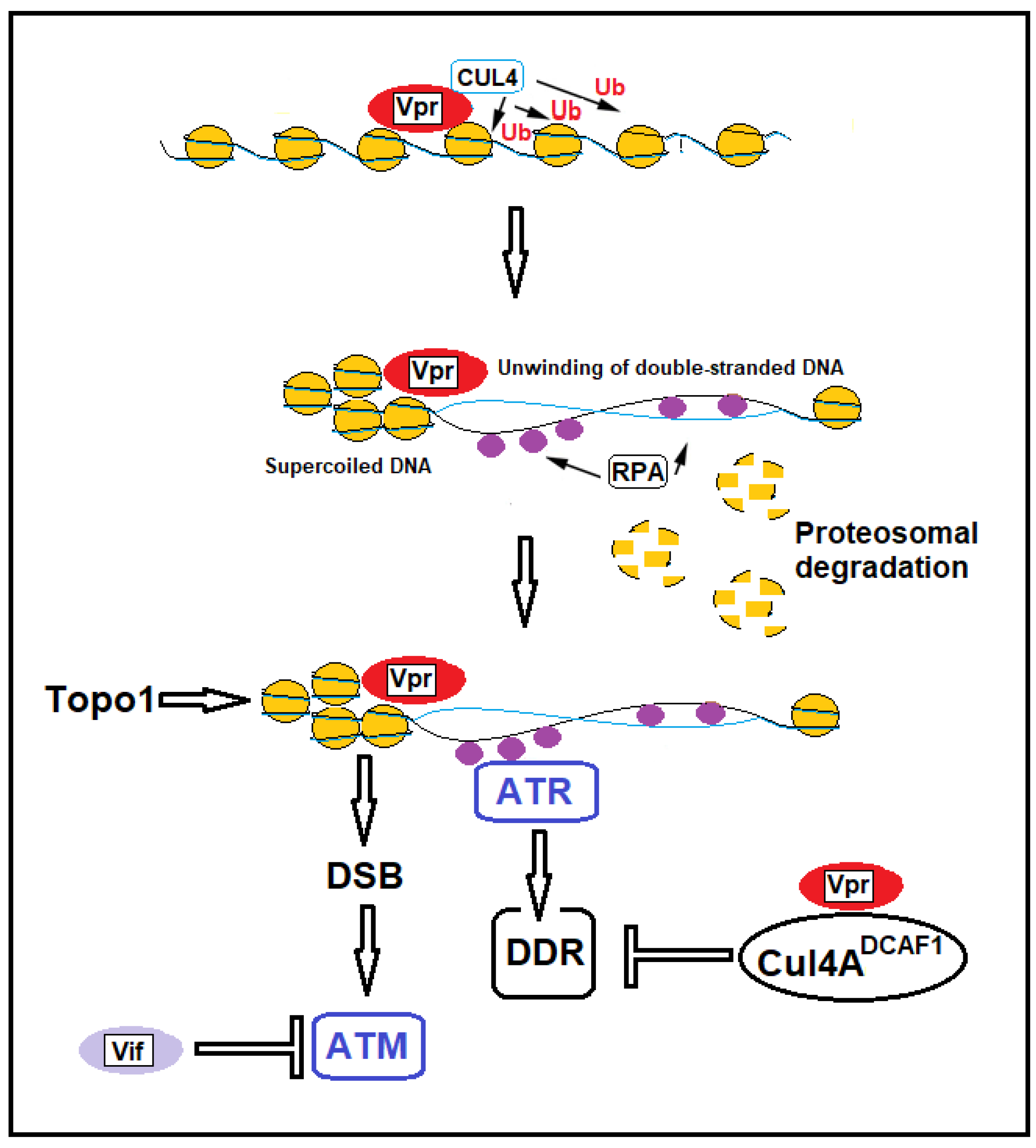

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




