Submitted:
27 March 2025
Posted:
29 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
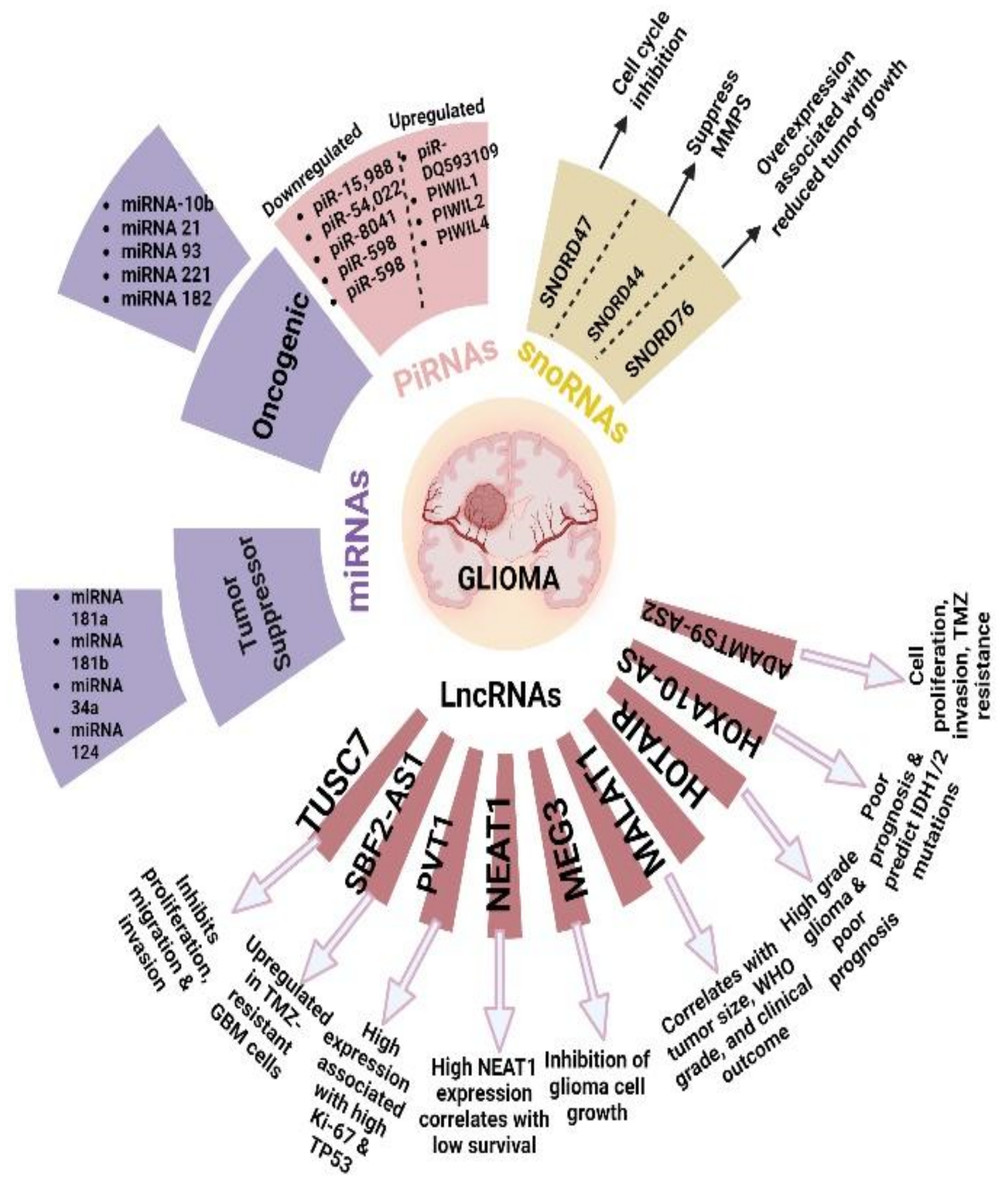
2.2. MicroRNAs
2.3. MicroRNAs as Biomarkers of GBM
2.4. MicroRNA-100 as a Tumor Biomarker of GBM
2.5. How miRNA Expression May Be Associated with Carcinogenesis
2.6. MicroRNA-504
2.7. MicroRNA-203
2.8. MicroRNA-301a
2.9. MicroRNA-145-5b
2.10. miR-29b
2.11. miR-451a, miR-485-3p, and miR-4298
2.12. miR-223 and miR-320e
2.13. miR-4763-3p, miR-1915-3p, and miR-3679-5p
2.14. Long Non-coding RNAs
2.15. HOTAIR
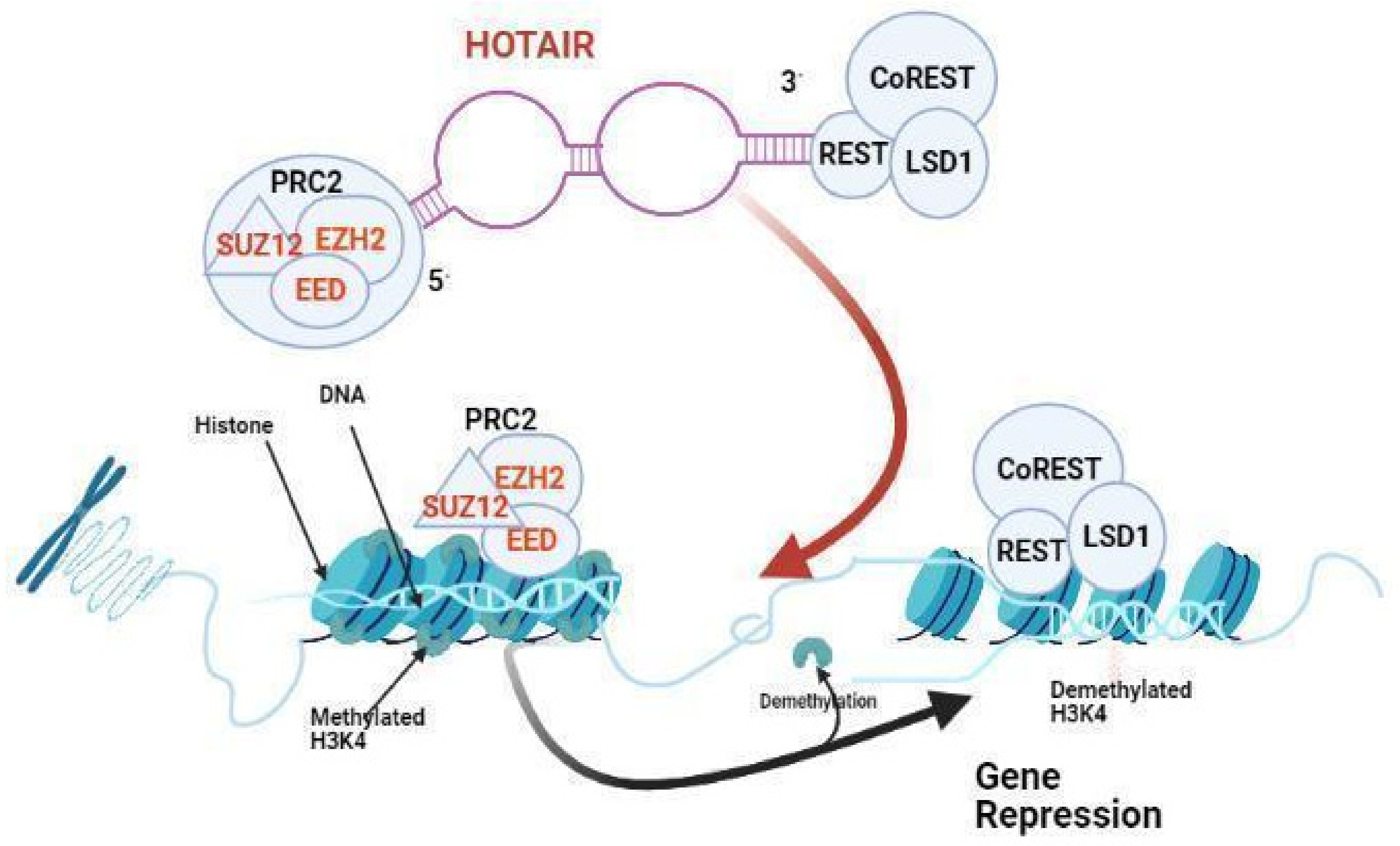
2.16. HOTAIR Expression Correlates with Glioma Grade
2.17. The Prognostic Significance of HOTAIR Expression in Glioma
2.18. HOTAIR as a Biomarker for Responsiveness of GBM Cells to BET Inhibitors
2.19. GBM Evolved Drug-Resistant Mechanisms by lncRNA SBF2-AS1
2.20. HOX-Related LncRNAs
2.21. MALAT1
2.22. H19
2.23. PVT1
2.24. NEAT1
2.25. Circular RNAs
2.26. PIWI-Interacting RNAs (piRNAs)
2.27. Small Nucleolar RNAs
3. Discussion
3.1. Different Challenges Facing Non-coding RNAs as Biomarkers for GBM
Challenges intrinsic to the RNA biological features
3.2. Sensitivity and Specificity
4. Conclusions and Future Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Hazra R, Debnath R, Tuppad A. Glioblastoma stem cell long non-coding RNAs: therapeutic perspectives and opportunities. Front Genet. 2024 Jul 2;15:1416772.
- Razavi, S.M., et al., Immune Evasion Strategies of Glioblastoma. Front Surg, 2016. 3: p. 11.
- Stupp, R., et al., Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med, 2005. 352(10): p. 987-96.
- Wu, L. and X. Qu, Cancer biomarker detection: recent achievements and challenges. Chem Soc Rev, 2015. 44(10): p. 2963-97.
- Kapranov, P., et al., RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science, 2007. 316(5830): p. 1484-8.
- Wahlestedt, C., Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov, 2013. 12(6): p. 433-46.
- Ma, L., V.B. Bajic, and Z. Zhang, On the classification of long non-coding RNAs. RNA Biol, 2013. 10(6): p. 925-33.
- Gomes, A.Q., S. Nolasco, and H. Soares, Non-coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci, 2013. 14(8): p. 16010-39.
- Kondo, Y., K. Shinjo, and K. Katsushima, Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci, 2017. 108(10): p. 1927-1933.
- Mercer, T.R., M.E. Dinger, and J.S. Mattick, Long non-coding RNAs: insights into functions. Nat Rev Genet, 2009. 10(3): p. 155-9.
- Lu, T.X. and M.E. Rothenberg, MicroRNA. J Allergy Clin Immunol, 2018. 141(4): p. 1202-1207.
- Barciszewska, A.M., MicroRNAs as efficient biomarkers in high-grade gliomas. Folia Neuropathol, 2016. 54(4): p. 369-374.
- Ryan, B.M., A.I. Robles, and C.C. Harris, Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer, 2010. 10(6): p. 389-402.
- Stark, A., et al., Identification of Drosophila MicroRNA targets. PLoS Biol, 2003. 1(3): p. E60.
- Grimson, A., et al., MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell, 2007. 27(1): p. 91-105.
- Filipowicz, W., S.N. Bhattacharyya, and N. Sonenberg, Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet, 2008. 9(2): p. 102-14.
- Lu, J., et al., MicroRNA expression profiles classify human cancers. Nature, 2005. 435(7043): p. 834-8.
- Zhang, H., et al., Serum miR-100 is a potential biomarker for detection and outcome prediction of glioblastoma patients. Cancer Biomark, 2019. 24(1): p. 43-49.
- Dong, L., et al., miRNA microarray reveals specific expression in the peripheral blood of glioblastoma patients. Int J Oncol, 2014. 45(2): p. 746-56.
- Wu, J., L. Li, and C. Jiang, Identification and Evaluation of Serum MicroRNA-29 Family for Glioma Screening. Mol Neurobiol, 2015. 52(3): p. 1540-1546.
- Wei, X., et al., Serum MicroRNA-125b as a Potential Biomarker for Glioma Diagnosis. Mol Neurobiol, 2016. 53(1): p. 163-170.
- Lai, N.S., et al., Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br J Cancer, 2015. 112(7): p. 1241-6.
- Sun, J., et al., Serum microRNA-128 as a biomarker for diagnosis of glioma. Int J Clin Exp Med, 2015. 8(1): p. 456-63.
- Yue, X., et al., Downregulation of serum microRNA-205 as a potential diagnostic and prognostic biomarker for human glioma. J Neurosurg, 2016. 124(1): p. 122-8.
- Li, H.Y., et al., Circulating microRNA-137 is a potential biomarker for human glioblastoma. Eur Rev Med Pharmacol Sci, 2016. 20(17): p. 3599-604.
- Regazzo, G., et al., A restricted signature of serum miRNAs distinguishes glioblastoma from lower grade gliomas. J Exp Clin Cancer Res, 2016. 35(1): p. 124.
- Liu, J., et al., MicroRNA-100 is a potential molecular marker of non-small cell lung cancer and functions as a tumor suppressor by targeting polo-like kinase 1. BMC Cancer, 2012. 12: p. 519.
- Lee, C., et al., Polo-like kinase 1 inhibition kills glioblastoma multiforme brain tumor cells in part through loss of SOX2 and delays tumor progression in mice. Stem Cells, 2012. 30(6): p. 1064-75.
- Xu, K., P. Liu, and W. Wei, mTOR signaling in tumorigenesis. Biochim Biophys Acta, 2014. 1846(2): p. 638-54.
- Grundmann, S., et al., MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation, 2011. 123(9): p. 999-1009.
- Tsuchiya, K., et al., Differentiation between solitary brain metastasis and high-grade glioma by diffusion tensor imaging. Br J Radiol, 2005. 78(930): p. 533-7.
- Jin, Z., et al., Serum expression level of miR-504 can differentiate between glioblastoma multiforme and solitary brain metastasis of non-small cell lung carcinoma. J buon, 2017. 22(2): p. 474-480.
- Chen, J., L. Yang, and X. Wang, Reduced circulating microRNA-203 predicts poor prognosis for glioblastoma. Cancer Biomark, 2017. 20(4): p. 521-526.
- Lan, F., et al., Serum exosomal miR-301a as a potential diagnostic and prognostic biomarker for human glioma. Cell Oncol (Dordr), 2018. 41(1): p. 25-33.
- Zhang, Y., et al., Diagnostic and prognostic significance of serum miR-145-5p expression in glioblastoma. Int J Clin Exp Pathol, 2019. 12(7): p. 2536-2543.
- Zhong, F., T. Huang, and J. Leng, Serum miR-29b as a novel biomarker for glioblastoma diagnosis and prognosis. Int J Clin Exp Pathol, 2019. 12(11): p. 4106-4112.
- Morokoff, A., et al., Serum microRNA is a biomarker for post-operative monitoring in glioma. J Neurooncol, 2020. 149(3): p. 391-400.
- Ohno, M., et al., Assessment of the Diagnostic Utility of Serum MicroRNA Classification in Patients With Diffuse Glioma. JAMA Netw Open, 2019. 2(12): p. e1916953.
- Fujimoto, A., et al., Erratum: Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet, 2016. 48(6): p. 700.
- Schlosser, K., et al., Assessment of Circulating LncRNAs Under Physiologic and Pathologic Conditions in Humans Reveals Potential Limitations as Biomarkers. Sci Rep, 2016. 6: p. 36596.
- Isaev, K., et al., Pan-cancer analysis of non-coding transcripts reveals the prognostic onco-lncRNA HOXA10-AS in gliomas. Cell Rep, 2021. 37(3): p. 109873.
- Sun, W., et al., Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet, 2017. 216-217: p. 105-110.
- Kumarswamy, R., et al., Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res, 2014. 114(10): p. 1569-75.
- Yang, Y., et al., Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin Sci (Lond), 2015. 129(8): p. 675-85.
- Tong, Y.S., et al., Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer, 2015. 14: p. 3.
- Siri, G., et al., A comprehensive review of the role of lncRNAs in gastric cancer (GC) pathogenesis, immune regulation, and their clinical applications. Pathol Res Pract, 2023. 241: p. 154221.
- Zhang, J.X., et al., HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro Oncol, 2013. 15(12): p. 1595-603.
- Tan, S.K., et al., Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer, 2018. 17(1): p. 74.
- Gao, Y., et al., Long Non-Coding RNA HOXA-AS2 Regulates Malignant Glioma Behaviors and Vasculogenic Mimicry Formation via the MiR-373/EGFR Axis. Cell Physiol Biochem, 2018. 45(1): p. 131-147.
- Cantile, M., et al., Hyperexpression of HOXC13, located in the 12q13 chromosomal region, in well-differentiated and dedifferentiated human liposarcomas. Oncol Rep, 2013. 30(6): p. 2579-86.
- Croce, C.M., LINCing chromatin remodeling to metastasis. Nat Biotechnol, 2010. 28(9): p. 931-2.
- Zhang, Y., et al., Circulating long non-coding HOX transcript antisense intergenic ribonucleic acid in plasma as a potential biomarker for diagnosis of breast cancer. Thorac Cancer, 2016. 7(6): p. 627-632.
- Berrondo, C., et al., Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes. PLoS One, 2016. 11(1): p. e0147236.
- Zhou, X., et al., HOTAIR is a therapeutic target in glioblastoma. Oncotarget, 2015. 6(10): p. 8353-65.
- Zhang, J.X., et al., Unique genome-wide map of TCF4 and STAT3 targets using ChIP-seq reveals their association with new molecular subtypes of glioblastoma. Neuro Oncol, 2013. 15(3): p. 279-89.
- Verhaak, R.G., et al., Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell, 2010. 17(1): p. 98-110.
- Pastori, C., et al., The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci U S A, 2015. 112(27): p. 8326-31.
- Zhang, Z., et al., Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma. J Exp Clin Cancer Res, 2019. 38(1): p. 166.
- Bridges, C.B., Current Maps of the Location of the Mutant Genes of Drosophila Melanogaster. Proc Natl Acad Sci U S A, 1921. 7(4): p. 127-32.
- Quinonez, S.C. and J.W. Innis, Human HOX gene disorders. Mol Genet Metab, 2014. 111(1): p. 4-15.
- Tsuboi, M., et al., The transcription factor HOXB7 regulates ERK kinase activity and thereby stimulates the motility and invasiveness of pancreatic cancer cells. J Biol Chem, 2017. 292(43): p. 17681-17702.
- Wu, S.Y., et al., A miR-192-EGR1-HOXB9 regulatory network controls the angiogenic switch in cancer. Nat Commun, 2016. 7: p. 11169.
- Nagata, H., et al., Genome-wide screening of DNA methylation associated with lymph node metastasis in esophageal squamous cell carcinoma. Oncotarget, 2017. 8(23): p. 37740-37750.
- Sui, B.Q., et al., HOXB13 expression and promoter methylation as a candidate biomarker in gastric cancer. Oncol Lett, 2018. 15(6): p. 8833-8840.
- Li, B., Q. Huang, and G.H. Wei, The Role of HOX Transcription Factors in Cancer Predisposition and Progression. Cancers (Basel), 2019. 11(4).
- Wang, J., et al., LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta, 2018. 485: p. 229-233.
- Zhao, H., et al., HOX antisense lincRNA HOXA-AS2 is an apoptosis repressor in all trans retinoic acid treated NB4 promyelocytic leukemia cells. J Cell Biochem, 2013. 114(10): p. 2375-83.
- Wang, F., et al., HOX Antisense lincRNA HOXA-AS2 Promotes Tumorigenesis of Hepatocellular Carcinoma. Cell Physiol Biochem, 2016. 40(1-2): p. 287-296.
- Zheng, F.X., et al., Long noncoding RNA HOXA-AS2 promotes cell migration and invasion via upregulating IGF-2 in non-small cell lung cancer as an oncogene. Eur Rev Med Pharmacol Sci, 2019. 23(11): p. 4793-4799.
- Xie, M., et al., Long noncoding RNA HOXA-AS2 promotes gastric cancer proliferation by epigenetically silencing P21/PLK3/DDIT3 expression. Oncotarget, 2015. 6(32): p. 33587-601.
- Shen, Z.H., K.M. Zhao, and T. Du, HOXA10 promotes nasopharyngeal carcinoma cell proliferation and invasion via inducing the expression of ZIC2. Eur Rev Med Pharmacol Sci, 2017. 21(5): p. 945-952.
- Eoh, K.J., et al., Dysregulated expression of homeobox family genes may influence survival outcomes of patients with epithelial ovarian cancer: analysis of data from The Cancer Genome Atlas. Oncotarget, 2017. 8(41): p. 70579-70585.
- Lim, J.Y., et al., Overexpression of miR-196b and HOXA10 characterize a poor-prognosis gastric cancer subtype. World J Gastroenterol, 2013. 19(41): p. 7078-88.
- Kurscheid, S., et al., Chromosome 7 gain and DNA hypermethylation at the HOXA10 locus are associated with expression of a stem cell related HOX-signature in glioblastoma. Genome Biol, 2015. 16(1): p. 16.
- Sun, Y. and L. Ma, New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers (Basel), 2019. 11(2).
- Kim, S.S., et al., Targeted nanocomplex carrying siRNA against MALAT1 sensitizes glioblastoma to temozolomide. Nucleic Acids Res, 2018. 46(3): p. 1424-1440.
- Huang, S.K., et al., A Panel of Serum Noncoding RNAs for the Diagnosis and Monitoring of Response to Therapy in Patients with Breast Cancer. Med Sci Monit, 2018. 24: p. 2476-2488.
- Duan, W., et al., Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget, 2016. 7(48): p. 78850-78858.
- Zhou, Q., et al., lncRNAs as potential molecular biomarkers for the clinicopathology and prognosis of glioma: A systematic review and meta-analysis. Gene, 2018. 668: p. 77-86.
- Li, J., et al., Clinicopathological and prognostic significance of long noncoding RNA MALAT1 in human cancers: a review and meta-analysis. Cancer Cell Int, 2018. 18: p. 109.
- Cao, S., et al., Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am J Cancer Res, 2016. 6(11): p. 2561-2574.
- Chung, W.Y., et al., Chromosome 11p15.5 regional imprinting: comparative analysis of KIP2 and H19 in human tissues and Wilms’ tumors. Hum Mol Genet, 1996. 5(8): p. 1101-8.
- Zhou, X., et al., Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep, 2015. 5: p. 11516.
- Shen, L., et al., Prognostic evaluation of serum long non-coding RNA H19 for endoscopic keyhole surgery or craniotomy in glioma. Ann Clin Biochem, 2020. 57(5): p. 365-372.
- Wang, W., et al., PVT1 Promotes Cancer Progression via MicroRNAs. Front Oncol, 2019. 9: p. 609.
- Han, Y., et al., Knockdown of lncRNA PVT1 Inhibits Glioma Progression by Regulating miR-424 Expression. Oncol Res, 2019. 27(6): p. 681-690.
- Fang, J. and J. Huang, Clinical significance of the expression of long non-coding RNA PVT1 in glioma. Cancer Biomark, 2019. 24(4): p. 509-513.
- Zou, H., et al., lncRNAs PVT1 and HAR1A are prognosis biomarkers and indicate therapy outcome for diffuse glioma patients. Oncotarget, 2017. 8(45): p. 78767-78780.
- Xue, W., et al., PVT1 regulates the malignant behaviors of human glioma cells by targeting miR-190a-5p and miR-488-3p. Biochim Biophys Acta Mol Basis Dis, 2018. 1864(5 Pt A): p. 1783-1794.
- Chen, Y., et al., Long Non-coding RNA Expression Profiling Identifies a Four-Long Non-coding RNA Prognostic Signature for Isocitrate Dehydrogenase Mutant Glioma. Front Neurol, 2020. 11: p. 573264.
- You, J., et al., MicroRNA-449a inhibits cell growth in lung cancer and regulates long noncoding RNA nuclear enriched abundant transcript 1. Indian J Cancer, 2014. 51 Suppl 3: p. e77-81.
- Chakravarty, D., et al., The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun, 2014. 5: p. 5383.
- Choudhry, H., et al., Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene, 2015. 34(34): p. 4546.
- He, C., et al., Aberrant NEAT1 expression is associated with clinical outcome in high grade glioma patients. Apmis, 2016. 124(3): p. 169-74.
- Sanger, H.L., et al., Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A, 1976. 73(11): p. 3852-6.
- Goodall, G.J. and V.O. Wickramasinghe, RNA in cancer. Nat Rev Cancer, 2021. 21(1): p. 22-36.
- Wang, Z. and X. Lei, Identifying the sequence specificities of circRNA-binding proteins based on a capsule network architecture. BMC Bioinformatics, 2021. 22(1): p. 19.
- Razavi, Z.S., et al., Gynecologic cancers and non-coding RNAs: Epigenetic regulators with emerging roles. Crit Rev Oncol Hematol, 2021. 157: p. 103192.
- Khanipouyani, F., H. Akrami, and M.R. Fattahi, Circular RNAs as important players in human gastric cancer. Clin Transl Oncol, 2021. 23(1): p. 10-21.
- Zhang, X., et al., circRNA_0005529 facilitates growth and metastasis of gastric cancer via regulating miR-527/Sp1 axis. BMC Mol Cell Biol, 2021. 22(1): p. 6.
- Xing, Y., et al., Circular RNA circ-Foxo3 inhibits esophageal squamous cell cancer progression via the miR-23a/PTEN axis. J Cell Biochem, 2020. 121(3): p. 2595-2605.
- Liu, J., et al., Overexpression of circular RNA circ-CDC45 facilitates glioma cell progression by sponging miR-516b and miR-527 and predicts an adverse prognosis. J Cell Biochem, 2020. 121(1): p. 690-697.
- Cao, Q., et al., Circular METRN RNA hsa_circ_0037251 Promotes Glioma Progression by Sponging miR-1229-3p and Regulating mTOR Expression. Sci Rep, 2019. 9(1): p. 19791.
- Vagin, V.V., et al., A distinct small RNA pathway silences selfish genetic elements in the germline. Science, 2006. 313(5785): p. 320-4.
- Gainetdinov, I., et al., A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains piRNA Production in Most Animals. Mol Cell, 2018. 71(5): p. 775-790.e5.
- Grivna, S.T., et al., A novel class of small RNAs in mouse spermatogenic cells. Genes Dev, 2006. 20(13): p. 1709-14.
- Wang, X., et al., MiRNA-154-5p inhibits cell proliferation and metastasis by targeting PIWIL1 in glioblastoma. Brain Res, 2017. 1676: p. 69-76.
- Qu, A., et al., A serum piRNA signature as promising non-invasive diagnostic and prognostic biomarkers for colorectal cancer. Cancer Manag Res, 2019. 11: p. 3703-3720.
- Krishnan, P. and S. Damaraju, The Challenges and Opportunities in the Clinical Application of Noncoding RNAs: The Road Map for miRNAs and piRNAs in Cancer Diagnostics and Prognostics. Int J Genomics, 2018. 2018: p. 5848046.
- Jacobs, D.I., et al., PIWI-Interacting RNAs in Gliomagenesis: Evidence from Post-GWAS and Functional Analyses. Cancer Epidemiol Biomarkers Prev, 2016. 25(7): p. 1073-80.
- Jacobs, D.I., et al., piRNA-8041 is downregulated in human glioblastoma and suppresses tumor growth in vitro and in vivo. Oncotarget, 2018. 9(102): p. 37616-37626.
- Williams, G.T. and F. Farzaneh, Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer, 2012. 12(2): p. 84-8.
- Hoang, C. and A.R. Ferré-D’Amaré, Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell, 2001. 107(7): p. 929-39.
- Kiss, T., Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. Embo j, 2001. 20(14): p. 3617-22.
- Runte, M., et al., The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet, 2001. 10(23): p. 2687-700.
- Xia, X.R., et al., Effects of small nucleolar RNA SNORD44 on the proliferation, apoptosis and invasion of glioma cells. Histochem Cell Biol, 2020. 153(4): p. 257-269.
- Appaiah, H.N., et al., Persistent upregulation of U6:SNORD44 small RNA ratio in the serum of breast cancer patients. Breast Cancer Res, 2011. 13(5): p. R86.
- Dong, X.Y., et al., SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum Mol Genet, 2008. 17(7): p. 1031-42.
- Dong, X.Y., et al., Implication of snoRNA U50 in human breast cancer. J Genet Genomics, 2009. 36(8): p. 447-54.
- Okugawa, Y., et al., Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut, 2017. 66(1): p. 107-117.
- Xu, B., et al., SNORD47, a box C/D snoRNA, suppresses tumorigenesis in glioblastoma. Oncotarget, 2017. 8(27): p. 43953-43966.
- Chen, L., et al., SNORD76, a box C/D snoRNA, acts as a tumor suppressor in glioblastoma. Sci Rep, 2015. 5: p. 8588.
- Fanelli, G.N., et al., LONG-NONCODING RNAs in gastroesophageal cancers. Noncoding RNA Res, 2018. 3(4): p. 195-212.
- Statello, L., et al., Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol, 2021. 22(2): p. 96-118.
- Hombach, S. and M. Kretz, Non-coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol, 2016. 937: p. 3-17.
- Baldassarre, A., et al., Circulating microRNAs and Bioinformatics Tools to Discover Novel Diagnostic Biomarkers of Pediatric Diseases. Genes (Basel), 2017. 8(9).
- Iempridee, T., et al., Identification of reference genes for circulating long noncoding RNA analysis in serum of cervical cancer patients. FEBS Open Bio, 2018. 8(11): p. 1844-1854.
- Onódi, Z., et al., Isolation of High-Purity Extracellular Vesicles by the Combination of Iodixanol Density Gradient Ultracentrifugation and Bind-Elute Chromatography From Blood Plasma. Front Physiol, 2018. 9: p. 1479.
- Linares, R., et al., High-speed centrifugation induces aggregation of extracellular vesicles. J Extracell Vesicles, 2015. 4: p. 29509.
- Corso, G., et al., Reproducible and scalable purification of extracellular vesicles using combined bind-elute and size exclusion chromatography. Sci Rep, 2017. 7(1): p. 11561.
- Necsulea, A., et al., The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature, 2014. 505(7485): p. 635-40.
- Pepe, M.S., et al., Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst, 2008. 100(20): p. 1432-8.
- Corchete, L.A., et al., Systematic comparison and assessment of RNA-seq procedures for gene expression quantitative analysis. Sci Rep, 2020. 10(1): p. 19737.
- Auer, P.L. and R.W. Doerge, Statistical design and analysis of RNA sequencing data. Genetics, 2010. 185(2): p. 405-16.
- Ray, P., et al., Statistical evaluation of a biomarker. Anesthesiology, 2010. 112(4): p. 1023-40.
- Zhou, Q., et al., MicroRNAs as potential biomarkers for the diagnosis of glioma: A systematic review and meta-analysis. Cancer Sci, 2018. 109(9): p. 2651-2659.
- Yang, C., et al., Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer, 2013. 132(1): p. 116-27.
- Morlion, A., et al., Custom long non-coding RNA capture enhances detection sensitivity in different human sample types. RNA Biol, 2021. 18(sup1): p. 215-222.
- Chen, M., et al., A novel biosensor for the ultrasensitive detection of the lncRNA biomarker MALAT1 in non-small cell lung cancer. Sci Rep, 2021. 11(1): p. 3666.
- Lobato-Delgado, B., B. Priego-Torres, and D. Sanchez-Morillo, Combining Molecular, Imaging, and Clinical Data Analysis for Predicting Cancer Prognosis. Cancers (Basel), 2022. 14(13).
- Manterola, L., et al., A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol, 2014. 16(4): p. 520-7.
- Wang, Z.Q., et al., Low serum level of miR-485-3p predicts poor survival in patients with glioblastoma. PLoS One, 2017. 12(9): p. e0184969.
- Yan, Y., et al., Novel Function of lncRNA ADAMTS9-AS2 in Promoting Temozolomide Resistance in Glioblastoma via Upregulating the FUS/MDM2 Ubiquitination Axis. Front Cell Dev Biol, 2019. 7: p. 217.
- Xavier-Magalhães, A., et al., Effects of the functional HOTAIR rs920778 and rs12826786 genetic variants in glioma susceptibility and patient prognosis. J Neurooncol, 2017. 132(1): p. 27-34.
- Shen, J., et al., Serum HOTAIR and GAS5 levels as predictors of survival in patients with glioblastoma. Mol Carcinog, 2018. 57(1): p. 137-141.
- Li, J., et al., Epigenetic repression of long non-coding RNA MEG3 mediated by DNMT1 represses the p53 pathway in gliomas. Int J Oncol, 2016. 48(2): p. 723-33.
- Cai, H., et al., Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene, 2017. 36(3): p. 318-331.
- Cai, H., et al., The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget, 2015. 6(23): p. 19759-79.
- Shang, C., et al., Long Non-coding RNA TUSC7, a Target of miR-23b, Plays Tumor-Suppressing Roles in Human Gliomas. Front Cell Neurosci, 2016. 10: p. 235.
- He, Z., et al., FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in Glioma. J Exp Clin Cancer Res, 2019. 38(1): p. 65.
- Liu, X., et al., SRSF10 inhibits biogenesis of circ-ATXN1 to regulate glioma angiogenesis via miR-526b-3p/MMP2 pathway. J Exp Clin Cancer Res, 2020. 39(1): p. 121.
- Qu, Y., et al., Circular RNA circ_0079593 indicates a poor prognosis and facilitates cell growth and invasion by sponging miR-182 and miR-433 in glioma. J Cell Biochem, 2019. 120(10): p. 18005-18013.
- Chen, C., et al., Circular RNA Pleiotrophin promotes carcinogenesis in glioma via regulation of microRNA-122/SRY-box transcription factor 6 axis. Eur J Cancer Prev, 2020. 29(2): p. 165-173.
- Shen, S., et al., PIWIL1/piRNA-DQ593109 Regulates the Permeability of the Blood-Tumor Barrier via the MEG3/miR-330-5p/RUNX3 Axis. Mol Ther Nucleic Acids, 2018. 10: p. 412-425.
- Li, J., et al., High expression of PIWIL2 promotes tumor cell proliferation, migration and predicts a poor prognosis in glioma. Oncol Rep, 2017. 38(1): p. 183-192.
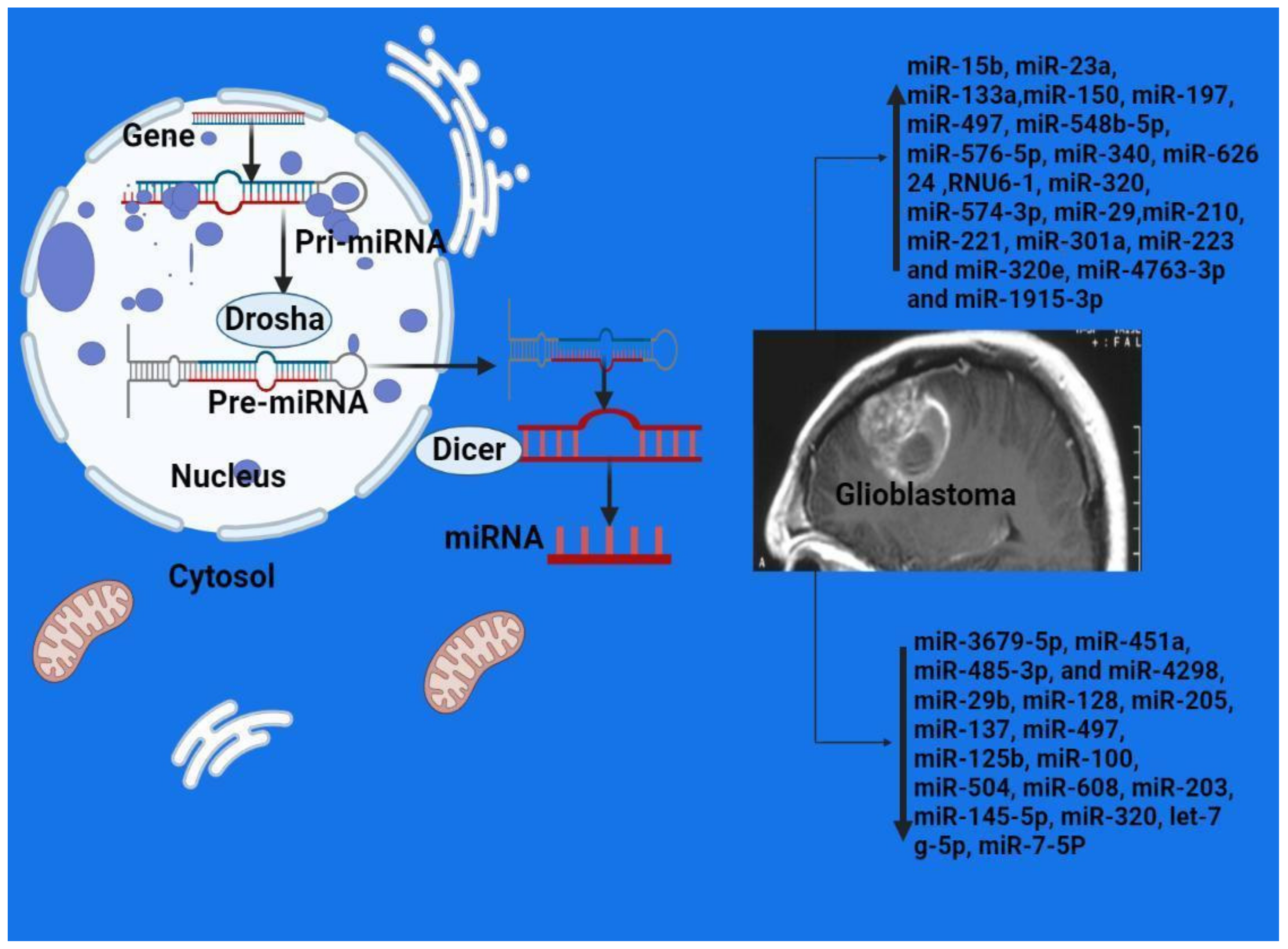
| Reference | Sample | Method | Overexpressed | Downexpressed |
|---|---|---|---|---|
| [18] 2013 |
GBM+ normal control | Microarray RT-PCR | miR-15b, miR-23a, miR-133a, miR-150, miR-197, miR-497, and miR-548b-5p | |
| [137] 2014 |
GBM+ normal control | Microarray RT-PCR | miR-576-5p, miR-340, and miR-626 24 | miR-320, let-7 g-5p, and miR-7-5P |
| [19] 2014 |
Glioma and normal controls | Microarray RT-PCR | RNU6-1, miR-320, and miR-574-3p | |
| [141] 2015 |
Glioma | RT-PCR | miR-29 | |
| [20] 2014 |
Glioma | RT-PCR | miR-125b | |
| [21] 2015 |
Glioma and normal controls | RT-PCR | miR-210 | |
| [22] 2015 |
Glioma and normal controls | RT-PCR | miR-128 | |
| [23] 2015 |
Glioma | RT-PCR | miR-205 | |
| [24] 2016 |
Glioma and normal controls | RT-PCR | miR-137 | |
| [25] 2016 |
Glioma and normal controls | RT-PCR | miR-497 and miR-125b | |
| [17] 2019 |
Glioma and normal controls | RT-PCR | miR-100 | |
| [26] 2017 |
Glioma and normal controls | RT-PCR | miR-221 | miR-504 and miR-608 |
| [32] 2017 |
Glioma and normal controls | RT-PCR | miR-203 | |
| [33] 2018 |
Glioma and normal controls | RT-PCR | miR-301a | |
| [34] 2019 |
Glioma and normal controls | RT-PCR | miR-145-5p | |
| [35] 2019 |
Glioma and normal controls | RT-PCR | miR-29b | |
| [36] 2017 |
Glioma and normal controls | RT-PCR | miR-451a, miR-485-3p, and miR-4298 | |
| [142] 2020 |
Glioma and normal controls | dd-PCR | miR-223 and miR-320e | |
| [37] 2020 |
Glioma and normal controls | RT-PCR | miR-4763-3p and miR-1915-3p | miR-3679-5p |
| Micro-RNA or/Targeted Micro-RNA | Official Title | Start Date | Study Type | Number Enrolled | Recruitment Status | Clinical Trial Number | Reference |
|---|---|---|---|---|---|---|---|
| Micro-RNAs level | Molecular Genetic, Host-derived and Clinical Determinants of Long-term Survival in Glioblastoma | July 5, 2015 | Observational | 599 | Active Non-recruiting | NCT03770468 | https://beta.clinicaltrials.gov/study/NCT03770468. accessed on 10 December 2022. |
| miRNA-10b | Evaluating the Expression Levels of MicroRNA-10b in Patients with Gliomas | May 2013 | Observational | 200 | Recruiting | NCT01849952 | https://www.clinicaltrials.gov/ct2/show/NCT01849952. accessed on 10 December 2022 |
| MicroRNAs level | Clinical Evaluation of a New Cancer Diagnosis Center at Kristianstad General Hospital, Sweden (CPF-DC) | October 2012 | Interventional (Clinical Trial) | 388 | Completed | NCT01709539 |
https://beta.clinicaltrials.gov/study/NCT01709539?tab=results# publications. accessed on 10 December 2022 |
| LncRNA Type | Cell Line | Method | Target Pathway |
Suggested Prognostic/Diagnostic Value | Reference |
|---|---|---|---|---|---|
| ADAMTS9-AS2 | T98G and U118 human glioma cell lines | qRT-PCR | mTOR pathway | Promotion of cell proliferation, migration and invasion Role in TMZ resistance | [143] |
| HOTAIR | U87MG and human glioma samples | qRT-PCR | Cell cycle and proliferation pathway | HOTAIR is overexpressed in GBM, it sustains cell proliferation. | [57] |
| HOTAIR | Serum exosomes | qRT-PCR | Cell cycle and proliferation pathway | Its expression was significantly correlated with high grade brain tumors. Its serum level from GBM patients was significantly higher than in the healthy volunteers. | [48] |
| HOTAIR | Human glioma tissue | qRT-PCR and RFLP | Cell cycle and proliferation pathway | HOTAIR SNPs rs920778 and rs12826786 do not play a significant role in glioma susceptibility and were associated with better survival of patients diagnosed with WHO grade III anaplastic oligodendroglioma. | [144] |
| HOTAIR | Patient serum | qRT-PCR | Cell cycle and proliferation pathway | High levels of HOTAIR were associated with higher mortality rates. | [145] |
| HOTAIR | Human glioma tissue and LN229 and U87 |
Gene set enrichment analysis |
Cell cycle and proliferation pathway | HOTAIR expression was related to higher glioma grade and poor prognosis. | [47] |
| HOXA10-AS | G432 and G797 and human glioma tissue | Machine learning | Unknown | HOXA10-AS activation is a robust marker of poor prognosis and can predict IDH1/2 mutations. | [41] |
| SBF2-AS1 | U87, LN229, A172, T98, U251, and (HEK) 293 T cells | qRT-PCR and FISH assays | Unknown | SBF2-AS1 was upregulated in TMZ-resistant GBM cells, and overexpression of SBF2-AS1 promoted resistance to TMZ. | [58] |
| MALAT1 | U87 and SHG139 and human glioma tissue | RT-qPCR | MAPK pathway | Expression of MALAT1 was significantly decreased in glioma specimens. MALAT1 expression was correlated with tumor size, WHO grade, and clinical outcome. | [80,81] |
| MEG3 | U251, U87 and A172 human glioma cell lines | RT-qPCR | p53 pathway PTEN pathway | Inhibition of glioma cell growth | [146] |
| PVT1 | Human glioma tissue | qRT-PCR | Cell cycle pathway BMP pathway NOTCH pathway | The expression levels of PVT1 were significantly higher in GBM tissues than in normal tissues, and PVT1 level was closely related to tumor grade. | [87] |
| PVT1 | Human glioma tissue | qRT-PCR | Cell cycle pathway, BMP pathway NOTCH pathway | High expression of PVT1 was associated with high Ki-67 level and more TP53 mutation and indicates poor survival. | [88] |
| PVT1 | U87 and U251 and human embryonic kidney cell line (HEK-293T) | qRT-PCR | Cell cycle pathway BMP pathway NOTCH pathway | PVT1 was upregulated in glioma specimens and cell lines. | [89] |
| NEAT1 | Human glioma tissue | qRT-PCR | WNT/β-Catenin pathway PI3K/AKT/mTOR pathway MEK/ERK pathway | Patients with high NEAT1 expression showed unfavorable overall survival. | [94] |
| TUG1 | Human glioma tissue and U251 MG, U87MG | qRT-PCR | WNT/β-Catenin pathway EMT pathway Neuronal differentiation pathway | Promotion of cell proliferation, migration and invasion regulation of glioma cell stemness enhances VEGF expression and angiogenesis | [147,148] |
| TUSC7 | Human glioma tissues and U251 , U87 | qRT-PCR | PI3K/Akt pathway | Inhibition of cell proliferation, migration and invasion | [149] |
| Authors | Type | Expression Level | Cell Line | Mechanism | Target Molecule | Reference |
|---|---|---|---|---|---|---|
| Cao et al. | hsa_circ_0037251 | Upregulated | U373, U251 (HEK) 293T cells | Inhibits apoptosis | miR-1229-3 mTORp | [103] |
| He et al., | circ_002136 | Upregulated | U87 | Promotes angiogenesis | miR-138-5p/SOX13 | [150] |
| Liu et a, | circ-ATXN1 | Upregulated | U87 | Promotes angiogenesis | miR-526b-3p/VEGFA | [151] |
| Qu et al., | circ_0079593 | Upregulated | U118, U251, U87MG, and LN229 | Prognostic marker | miR-182 | [152] |
| Chen et al., | circ_PTN | Upregulated | U251 and U87 | Promotes cellular proliferation | miR-122/SOX6 | [153] |
| piRNA type | Cell Line | Expression Level | Reference |
|---|---|---|---|
| piR-15,988 | Human glioma tissue | Downregulated | [111] |
| piR-54,022 | Human glioma tissue | Downregulated | [111] |
| piR-8041 | Human glioma tissue | Downregulated | [111] |
| piR-DQ593109 | U87 cell line and HEK293T | Upregulated | [154] |
| PIWIL1 | U87 cell line and HEK293T | Upregulated | [154] |
| piR-598 | U87 and A172 and human glioma tissue | Downregulated | [110] |
| PIWIL2 | H4, A172, U251, U87, and U118 and human glioma tissue | Upregulated | [155] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





