Submitted:
29 March 2025
Posted:
31 March 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Materials & Methods
Study Design and Population
Statistical Analysis
Results
Discussion
Conclusions
References
- Coronaviruses. National Foundation for Infectious Diseases. (2022). Accessed: 05/22: https://www.nfid.org/infectious-diseases/coronaviruses.
- World Health Organization. (2022). Accessed: 05/22: https://www.who.int/publications/m/item/covid-19-public-health-emergency-ofinternational- concern-(pheic)-global-rese.
- (By. Coronavirus world map: Tracking the global outbreak. The New York Times). Accessed: 2021282020: https://www.nytimes.com/interactive/2021/world/covid-cases.html.
- Velavan TP, Meyer CG: Mild versus severe covid- 19: Laboratory markers. International Journal of Infectious Diseases. 2020, 95:304-307. [CrossRef]
- Kilercik M, Demirelce Ö, Serdar MA, Mikailova P, Serteser M: A new haematocytometric index: Predicting severity and mortality risk value in COVID-19 patients. PLOS ONE. 2021, 16. [CrossRef]
- Tan L, Wang Q, Zhang D, et al.: Lymphopenia predicts disease severity of covid- 19: A descriptive and predictive study. Signal Transduction and Targeted Therapy. 2020, 5:10. [CrossRef]
- Linssen J, Ermens A, Berrevoets M, et al.: A novel haemocytometric covid-19 prognostic score developed and validated in an observational multicentre European Hospital-based study. eLife. 2020, 9. [CrossRef]
- Wang C, Deng R, Gou L, et al.: Preliminary study to identify severe from moderate cases of covid-19 using combined hematology parameters. Annals of Translational Medicine. 2020, 8:593-593. [CrossRef]
- Bellan M, Azzolina D, Hayden E, et al.: Simple parameters from complete blood count predict in-hospital mortality in COVID-19. 2021-2021. [CrossRef]
- Zheng Y, Zhang Y, Chi H, et al.: The hemocyte counts as a potential biomarker for predicting disease progression in covid- 19: A retrospective study. Clinical Chemistry and Laboratory Medicine (CCLM. 2020, 58:1106-1115. [CrossRef]
- Hu B, Huang S, Yin L: The cytokine storm and Covid-19. Journal of Medical Virology. 2020, 93:250-256. [CrossRef]
- Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R: The COVID-19 cytokine storm; what we know so far. Frontiers in Immunology. 2020, 11. [CrossRef]
- Tavakolpour S, Rakhshandehroo T, Wei EX, Rashidian M: Lymphopenia during the COVID-19 infection: What it shows and what can be learned. Immunology Letters. 2020, 225:31-32. [CrossRef]
- Diao B, Wang C, Tan Y, et al.: Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Frontiers in Immunology. 2020, 11. [CrossRef]
- Xiang Q, Feng Z, Diao B, et al.: SARS-COV-2 induces lymphocytopenia by promoting inflammation and decimates secondary lymphoid organs. Frontiers in Immunology. 2021, 12. [CrossRef]
- Cavaillon J-M, Adib-Conquy M: Immune status in sepsis: The bug, the site of infection and the severity can make the difference. Critical Care. 2010, 14:167. [CrossRef]
- Drewry AM, Samra N, Skrupky LP, Fuller BM, Compton SM, Hotchkiss RS: Persistent lymphopenia after diagnosis of sepsis predicts mortality. Shock. 2014, 42:383-391. [CrossRef]
- Meyer NJ, Lindell RB, Wherry EJ: Immune stimulation with recombinant human granulocyte colony-stimulating factor for coronavirus disease 2019 (covid-19)—beware of blind spots. JAMA Internal Medicine. 2021, 181:78. [CrossRef]
- Huang C, Wang Y, Li X, et al.: Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The. Lancet. 2020, 395:497-506. [CrossRef]
| Variables | Hospital Mortality | P value | |
| No (Survivors) | Yes (Non-survivors) | ||
| N (%) | 3461 (88%) | 464 (12%) | |
| Patient characteristics | |||
| Age, years, median (IQR) | 67 (54-79) | 71 (61-81) | < .0001 |
| Sex, n (%) | |||
| Male | 1838 (53%) | 309 (67%) | < .0001 |
| Female | 1623 (47%) | 155 (33%) | < .0001 |
| Race, n (%) | |||
| White | 2707 (78%) | 363 (78%) | ns |
| Black | 341 (10%) | 41 (9%) | ns |
| Other | 413 (12%) | 60 (13%) | ns |
| BMI, kg/m2, median (IQR) | 28.7 (24.6-34.2) | 30.8 (25.9-36.0) | < .0001 |
| Elixhauser comorbidities, median (IQR) | 3 (2-5) | 5 (3-6) | < .0001 |
| Top 9 comorbidity categories, n (%) | |||
| Hypertension (pooled) | 2337 (68%) | 336 (72%) | ns |
| Obesity | 1157 (33%) | 220 (47%) | < .0001 |
| Neurologic diseases (pooled) | 602 (17%) | 215 (46%) | < .0001 |
| Diabetes mellitus (pooled) | 1031 (30%) | 172 (37%) | .0014 |
| Coagulopathy | 500 (14%) | 159 (34%) | < .0001 |
| Renal failure (pooled) | 571 (16%) | 132 (28%) | < .0001 |
| Chronic pulmonary disease | 752 (22%) | 106 (23%) | ns |
| Heart failure (pooled) | 474 (14%) | 105 (23%) | < .0001 |
| Iron deficiency anemia | 690 (20%) | 181 (39%) | < .0001 |
| CBC parameters and ratios, median (IQR) | |||
| WBC, 103 cells/mL | 6.9 (5.1-9.7) | 7.6 (5.0-11.1) | .0025 |
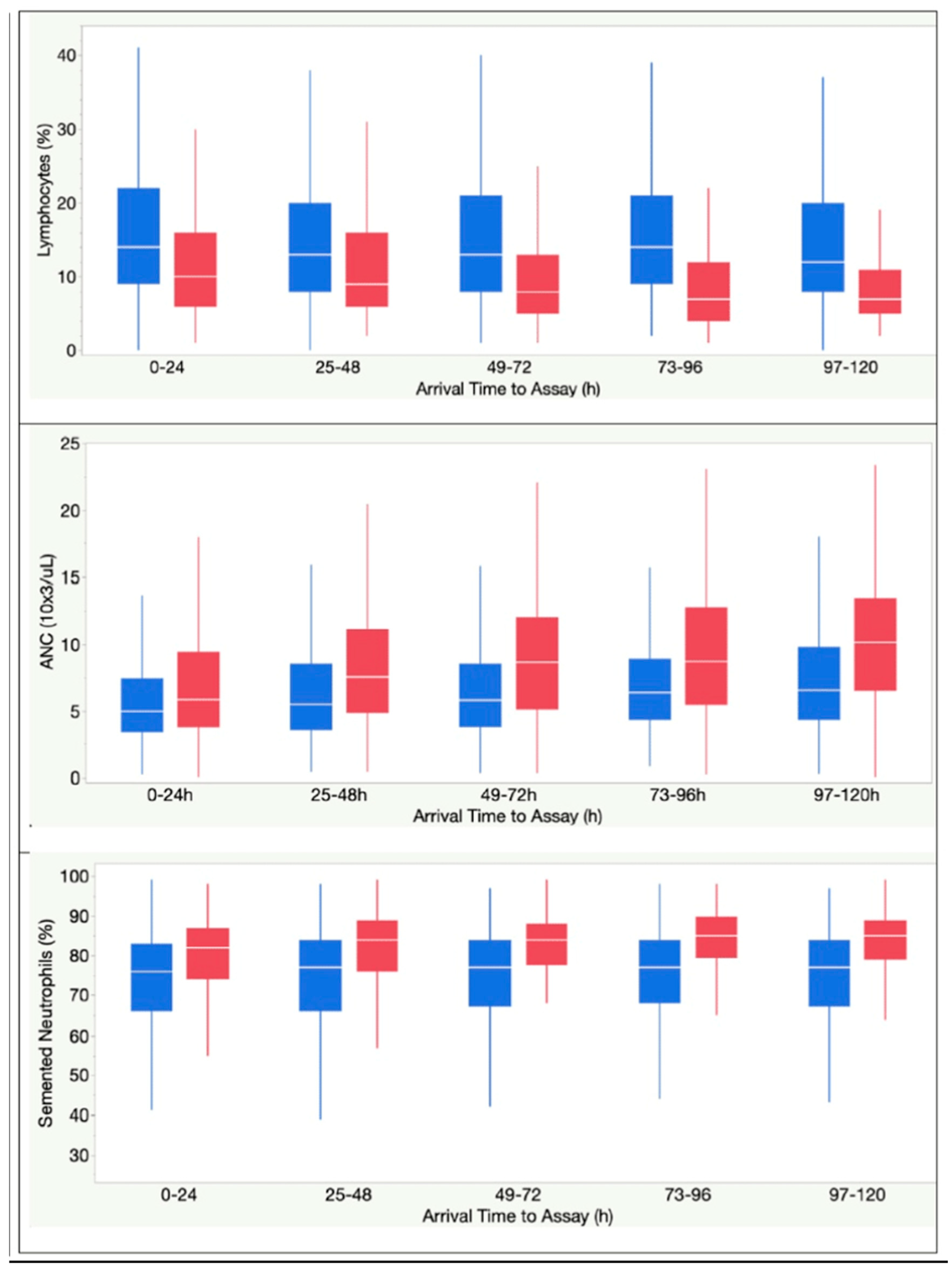
| Segmented neutrophils, % | 75 (66-82) | 81 (74-87) | < .0001 |
| Lymphocytes, % | 14 (9-21) | 10 (5-15) | < .0001 |
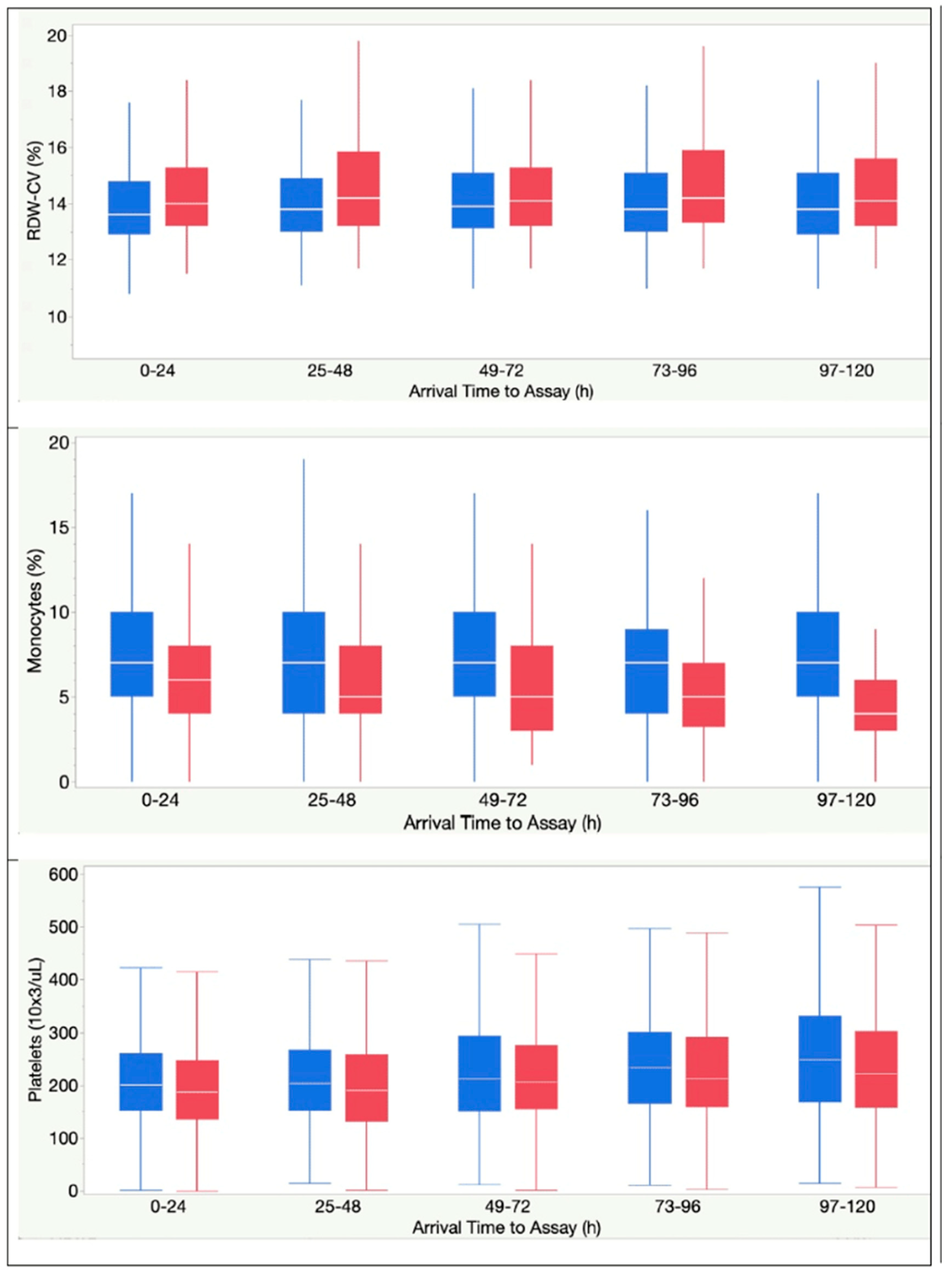
| Monocytes, % | 8 (5-10) | 4 (3-7) | < .0001 |
| Eosinophils, % | 0 (0-1) | 0 (0-0) | ns |
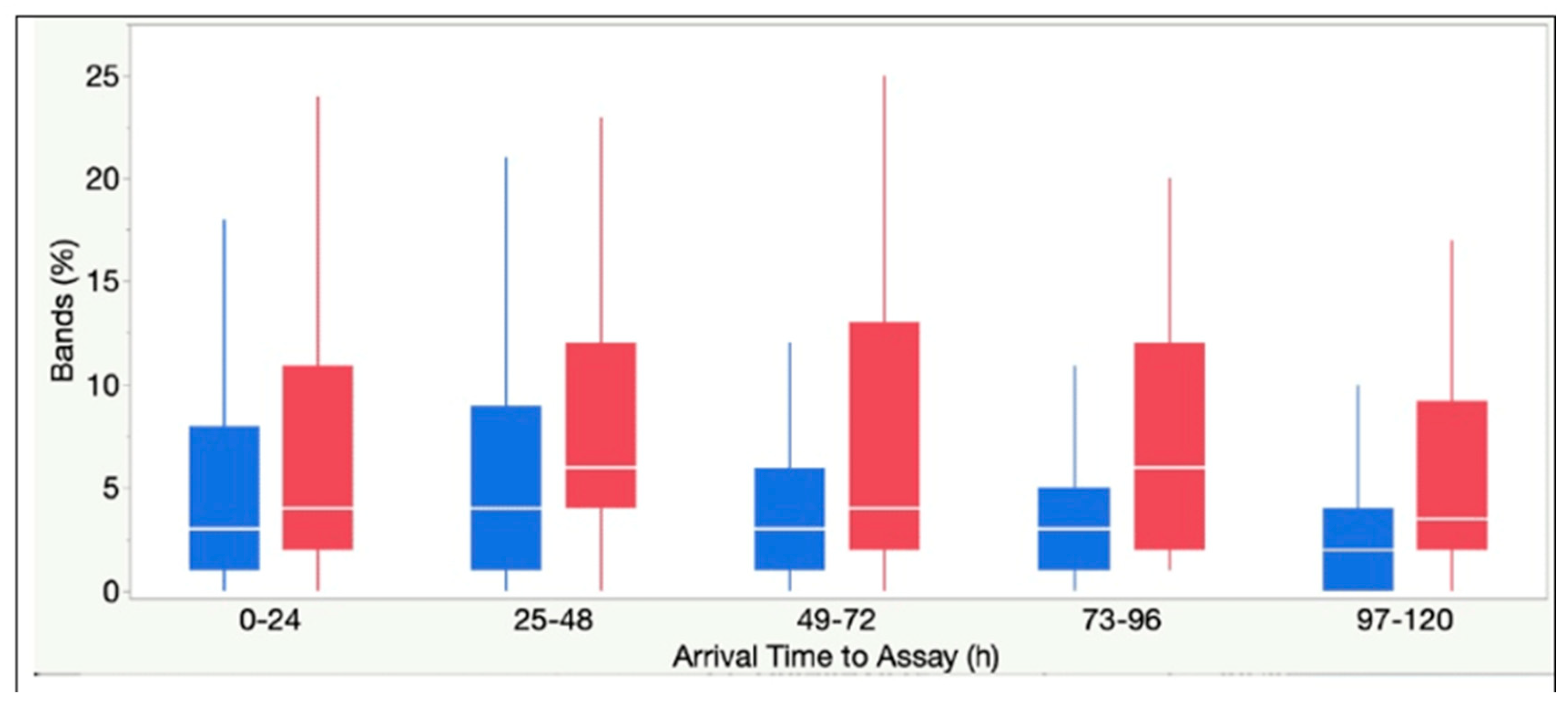
| Bands, % | 3 (1-7) | 4 (2-12) | < .0001 |
| Absolute Neutrophil Count, 103 ells/mL | 5.10 (3.51-7.54) | 5.97 (3.85-9.28) | < .0001 |
| Hemoglobin, gm/dL | 13.2 (11.7-14.5) | 13.3 (11.7-14.7) | ns |
| RDW-SD, (fL) | 44.5 (41.4-48.5) | 45.6 (42.8-50.3) | ns |
| RDW-CV, % | 13.6 (12.9-14.7) | 14.0 (13.2-15.2) | < .0001 |
| Platelet, 103 cells/mL | 205 (158-268) | 190 (142-257) | .0001 |
| ANC/ALC ratio | 5.5 (3.2-9.4) | 8.3 (5.0-16.4) | < .0001 |
| APC/ALC ratio | 222 (145-333) | 273 168-444) | < .0001 |
| Inflammatory markers, median (IQR) | |||
| CRP, mg/dL | 6.9 (2.8-12.1) | 11.4 (6.9-17.4) | < .0001 |
| Ferritin, ng/mL | 456 (203-935) | 966 (494-1709) | < .0001 |
| LDH, U/L | 308 (233-415) | 458 (355-648) | < .0001 |
| Coagulation markers, median (IQR) | |||
| D-dimer, mg/mL | 0.90 (0.54-1.71) | 1.30 (0.79-3.55) | < .0001 |
| Pro Time, s | 11.3 (10.8-12.0) | 11.5 10.9-12.8) | < .0001 |
| INR | 1.07 (1.02-1.14) | 1.09 (1.03-1.22) | < .0001 |
| Laboratory features in inflammatory biomarkers model (2a) | Percentage of feature contribution | Percentage of feature contribution | Overall R2 of model |
| LDH (U/L) | 45% | 53% | |
| CRP (mg/dL) | 30% | ||
| Ferritin (ng/mL) | 26% | ||
| Laboratory features in CBC model (2b) | Percentage of feature contribution | Percentage of feature contribution | Overall R2 of model |
| Monocytes (%) | 16% | 63% | |
| Lymphocytes (%) | 15% | ||
| Platelet (10X3/UL) | 15% | ||
| ANC (10X3/UL) | 15% | ||
| RDW-CV (%) | 15% | ||
| Segmented Neutrophils (%) | 13% | ||
| Bands (%) | 10% | ||
| Laboratory features in combined model (2c) | Percentage of feature contribution | Percentage of feature contribution | Overall R2 of model |
| LDH (U/L) | 45% | 69% | |
| RDW-CV (%) | 11% | ||
| Monocytes (%) | 11% | ||
| Platelet (10X3/UL) | 9% | ||
| Lymphocytes (%) | 9% | ||
| ANC (10X3/UL) | 8% | ||
| CRP (mg/dL) | 8% | ||
| Ferritin (ng/mL) | 8% | ||
| Segmented Neutrophils (%) | 8% | ||
| Bands (%) | 7% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




