1. Introduction
Leaf rust caused by
Puccinia triticina Eriks is one of the most devastating diseases affecting common wheat (
Triticum aestivum L.) worldwide [
1]. The leaf rust pathogen primarily attacks the leaf blades, although it can also infect the leaf sheath and glumes in highly susceptible cultivars. This disease can cause yield loss of up to 40% under epidemic conditions, mainly through reductions in kernel weight and kernel numbers per spike [
2]. Although leaf rust can be controlled by fungicides, the use of resistant cultivars is more effective, economic, and environmentally safe [
3].
Resistance to leaf rust is broadly classified into two race specific and race non-specific resistance. Race-specific resistance is effective against some but not all pathogen pathotypes, and it tends to involve one or more genes that elicit a hypersensitive response to avirulent pathotypes[
4]. This type of resistance predominantly applies throughout the entire growth cycle and therefore has been described as “all-stage resistance.” Race-non-specific resistance is quantitatively inherited, usually manifests at later growth stages, and is often effective against multiple pathogens[
5]. This type of resistance is also referred to as adult plant resistance (APR) or slow-rusting resistance. The important feature of race non-specific resistance is its durability[
6]. Currently 83 leaf rust resistance genes in wheat have been mapped to chromosome locations and assigned gene designations[
7]. Although most of the reported genes confer race-specific resistance, only a few confer race nonspecific resistance. Identification of new slow-rusting genes is important for breeding cultivars with durable resistance.
Molecular markers including restriction fragment length polymorphisms (RFLP), amplified fragment length polymorphisms (AFLP), simple sequence repeats (SSR), resistance gene analogue polymorphisms (RGAP), and single nucleotide polymorphisms (SNP), have been widely used in the genetic mapping of wheat disease resistance genes. Among them, high-density SNP arrays provide a superior approach for QTL mapping due to their higher accuracy and higher density compared to other markers[
8,
9,
10].
The Italian common wheat cultivar N. Strampelli, introduced to the Gansu Province of China in 1973 exhibits adult plant resistance to stripe rust that is controlled by three QTLs including
Lr18[
11]. For example, observations of cultivar N. Strampelli in Longnan, Gansu, China showed that this cultivar was generally immune or highly resistant to leaf rust, and the disease resistance of this cultivar was stable despite environmental changes for over 30 years.
The objective of this study was to explore the genetic basis of durable leaf rust resistance in cultivar N. Strampelli. To further analyze adult leaf rust resistance genes in this cultivar and to locate the QTLs associated with leaf rust resistance, 193 RIL populations of N. Strampelli×Huixianhong were monitored in Baoding, Hebei Province, China, and Zhoukou, Henan Province, China, from 2020 to 2022. QTLs associated with resistance were identified based on phenotypic characters and SNP/SSR genotyping. The objectives of this study were to detect QTLs associated with the APR in N. Strampelli.
2. Materials and Methods
Plant materials and Puccinia triticina pathotypes
The N. Strampelli×Huixianhong population consisted of 193 RILs. The cultivar Zhengzhou 5389 was used as the susceptible control.
Puccinia triticina races in the experimental plots
To obtain some information on number and virulence of Pt races in the experimental plots, race identification was done on samples from the experiment in Hebei in 2020. Briefly, Zhengzhou 5389 leaves bearing uredinia were collected from the experimental plots. Single uredinia were isolated, and then the spores propagated. The resulting urediniospores were used to inoculate the wheat host differentials that consisted of 16 nearby isogenic lines, with an additional fourth letter name. Four Pt pathotypes (THTS, THTQ, PHPS, and THTT) were used in the field.
Field
Leaf rust severities of wheat RIL populations field trials were conducted in Baoding, Hebei province, China (115.47°E, 38.85°N) and in Zhoukou, Henan province, China (114.53°E, 33.80°N). Three autumn-sown wheat crop seasons data were collected during the 2019/2020, 2020/2021, and 2021/2022 cropping seasons in Baoding and during the 2020/2021 cropping season in Zhoukou. The corresponding datasets are referred to herein as 2020BD, 2021BD, 2022BD, and2020HN, respectively. The experiments were conducted for N. Strampelli×Huixianhong (193 RILs) in Hebei and Henan in 2020 and 2022. Each of the four location × year combinations was considered as an individual environment. In each environment, the experiment was in a randomized complete block design with two replicates. An individual plot consisted of a single-e1-m-long row with 50cm between adjacent rows. Each plot was sown with approximately 20 seeds of a RIL. A control wheat Zhengzhou 5389 was included after every 10 RIL rows. Two rows of Zhengzhou 5389 perpendicular and adjacent to the plot rows were sown along each field block to serve as spreader plants of inoculums. Leaf rust infection was initiated by spraying the spreader at tillering stage with a water suspension of an equal urediniospores mixture of Pt pathotypes THTS, THTQ, PHPS, and THTT to which a few drops of Tween 20 (0.03%) were added.
Disease severity was scored based on the percentage of leaf area covered with uredinia according to the modified Cobb scale and was assessed three times per cropping season at about 1 week intervals, with the first scoring performed 4 weeks after inoculation in each environment.
Statistical analysis
Significant differences in phenotype and final disease severity (FDS) among trials (2020BD, 2021BD, 2022BD, and 2020HN) were identified using analysis of variance (ANOVA) in Microsoft Excel.
Molecular markers
Genomic DNA samples were extracted from the leaves of uninfected seedlings (both parental lines and RILs) using the CTAB method[
12]. The 193 RILs and the parental lines were genotyped using Affymetrix 660K SNP arrays (660,009 markers) by Capital Bio Technology Company (Beijing, China). Monomorphic SNPs (heterozygosity > 25%), those with high deletion rates (> 25% missing data), and those with distorted segregation ratios (minimum gene frequency > 5%) were removed. The remaining high-quality markers were used for SNP analysis. Genotypic data associated with 21 additional SSR markers were also collected[
13].
PCR reaction system consisted of 1.0μl 10×PCR buffer (Mg2+), 5μL 2×Taq PCR Master Mix, 1.0μl forward primer (10 molμL−1), 1.0μl reverse primer (10 molμL−1), 2.0μl DNA template (50 ng/μl), and 3.0μl ddH2O. While the normal PCR cycling was performed for most of the markers, for certain markers, a touchdown program was used, which started with 94°C for 5 min; then 35 cycles of 94°C denaturing for 30 s, 60°C annealing for 30 s (touchdown with 0.5°C each cycle), and 72°C extending for 30 s; followed by35 cycles of 94°C for 30 s, 55°C for 30 s, and72°C for 30 s; and ended with 72°C for 10 min. Amplicons were separated in 12% denaturing polyacrylamide gels and visualized with the silver staining method. Linkage map was constructed using Map Manager QTXb2.0 and QTL Icimapping 4.0.
Genotyping,map construction and QTL analysis
Mapping analyses to detect QTLs associated with leaf rust FDS in each trial was performed using the Inclusive Composite Interval Mapping (ICIM) algorithm in IciMapping 4.1[
14]. QTL detection was performed using the composite inclusive Composite Interval Mapping (ICIM),The threshold LOD score was calculated by running the permutation program set with 1000 replicates at a type I error rate of α=0.05. Permutation analysis was conducted for the experiments , the highest threshold LOD value (2.5) was chosen as a uniform threshold for all experiments. Effect size of QTL was measured with determination coefficient.
Genotypic data were filtered to retain only markers with missing values <5% RILs with missing values >5% were filtered out. After removing redundant markers identified applying the Bin function in QTL IciMapping V4.1 software, the retained high quality markers (each marker representing a unique locus) were grouped and ordered using JoinMap 4.0. Genetic distances (in cM) were estimated based on the Kosambi mapping function. A linkage group was split if there was a distance ≥20 cM between two adjacent markers.
Stepwise regressions were used to detect the percentage of variance explained (PVE) by individual QTLs and additive effects at LOD peaks. Linkage groups were then orientated and assigned to chromosomes through alignments of the sequences flanking SSR with chromosomal survey sequence map. The sequences flanking all SNP probes were searched against the wheat reference sequence (cultivar Chinese Spring;
https://urgi.versailles.inra.fr/blast_iwgsc/blast.php, IWGSC2022) using BLAST to determine physical positions.Factorial ANOVAs were conducted to determine the significance of interactions between stably detected resistance loci based on FDS.
3. Results
Resistance of N. Strampelli×Huixianhong RILs to leaf rust
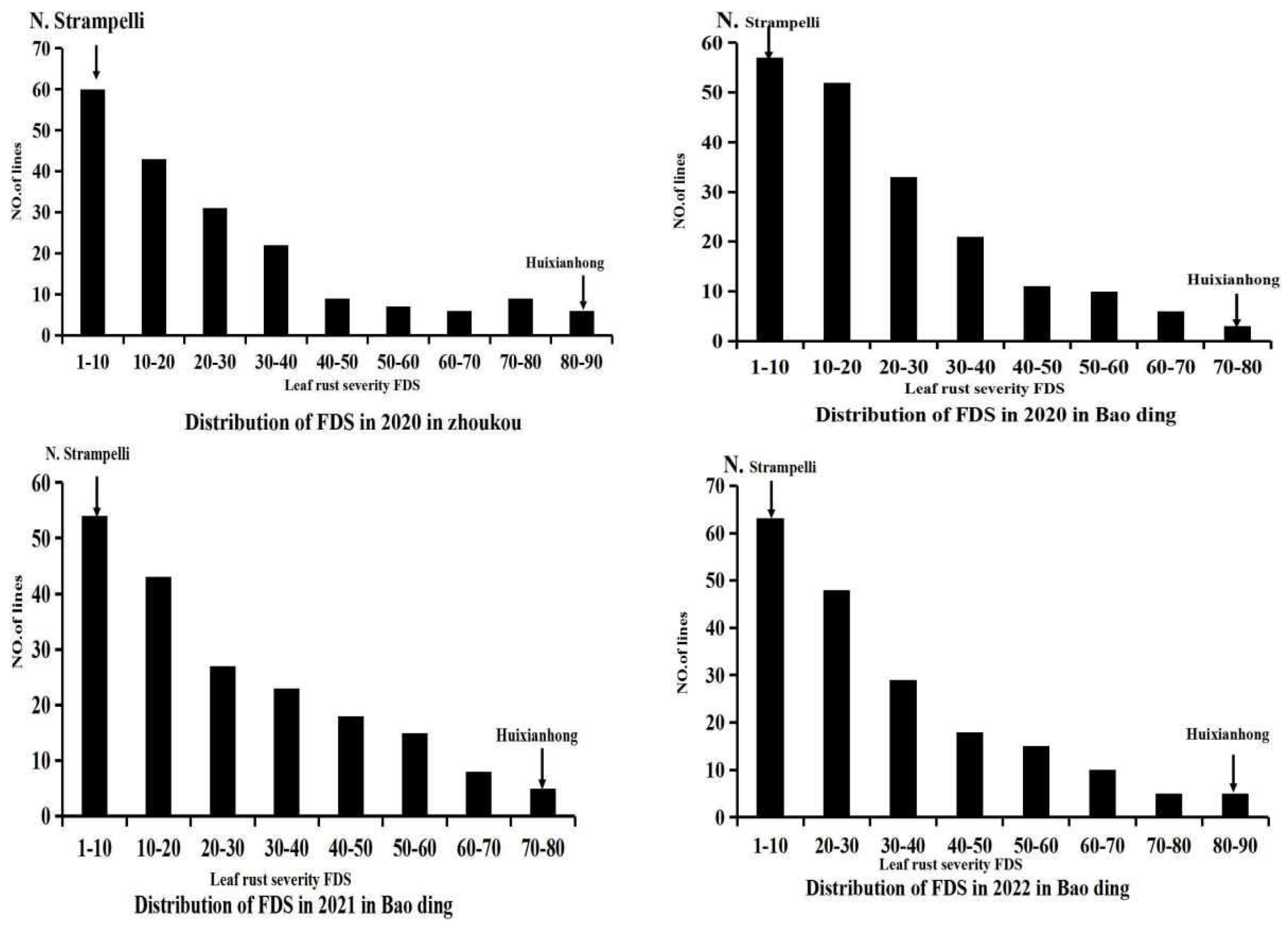
In each of the four field experiments, the leaf rust severities of susceptible control Zhengzhou 5389 ranged from 90%to 100%, leaf rust severities were approximately 80% on the susceptible control wheat Huixianhong and less than 10% on the resistant landrace N. Strampelli as exemplified by the typical symptoms shown in
Figure 1. This indicates that disease pressure was adequately high for revealing resistance and susceptibility. This suggested that both parents possess minor resistance genes. Mean leaf rust FDS for the RILs was 30.5-45% across all trials (
Table 1). The frequency distribution of leaf rust severities in each trial showed a continuous distribution skewed towards resistance (
Figure 1), indicating polygenic inheritance. Pearson correlation comparisons showed that FDS was significantly correlated between all pairs of trials, with Pearson correlation coefficients (R) of 0.7-0.8 (p<0.05;
Table 2). Correlation coefficients for among the experiments were all significant with p<0.05, ranging from 0.7 to 0.8 in N. Strampelli×Huixianhong. Variances due to RIL and environment were significant and predominated though the interaction variances were also significant. ANOVAs confirmed that there was significant variation among the genotypes Ril×Environment.
SNP and SSR genotypes
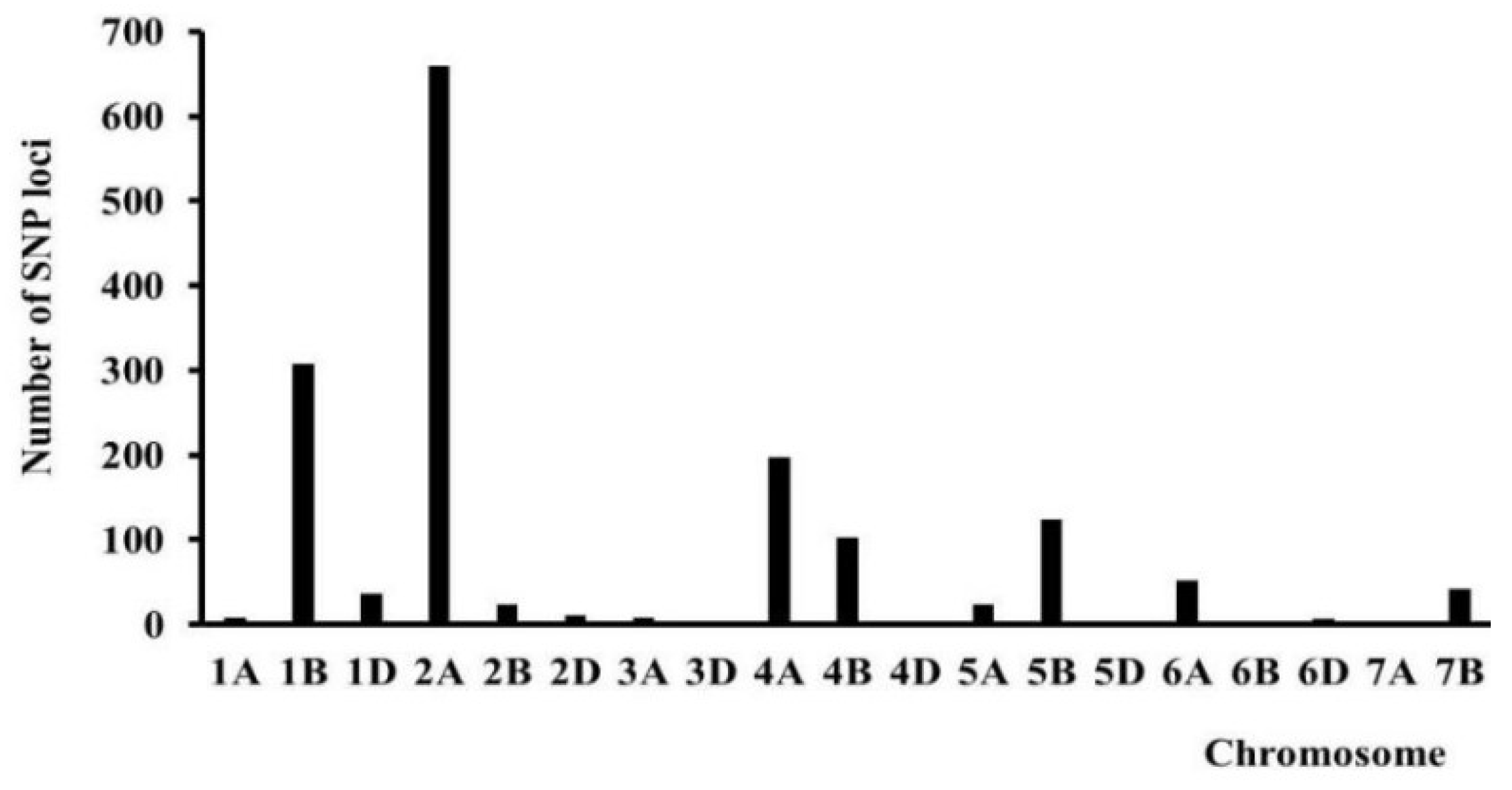
The distribution of SNP markers across chromosomes, in combination with its histogram, showed that the polymorphic loci controlling the FDS-associated QTLs were mainly distributed on chromosomes 1B, 2A, 5B, and 7D (
Figure 2). After inoculation with four
Pt pathotypes, 1200 SSR markers were identified between the parental lines and the bulked RILs. Three QTLs were identified on chromosomes 2A (
QLr.hbau-2A.1 and
QLr.hbau-2A.2), and 5B (
QLr.hbau-5B).
QTL mapping for leaf rust resistance
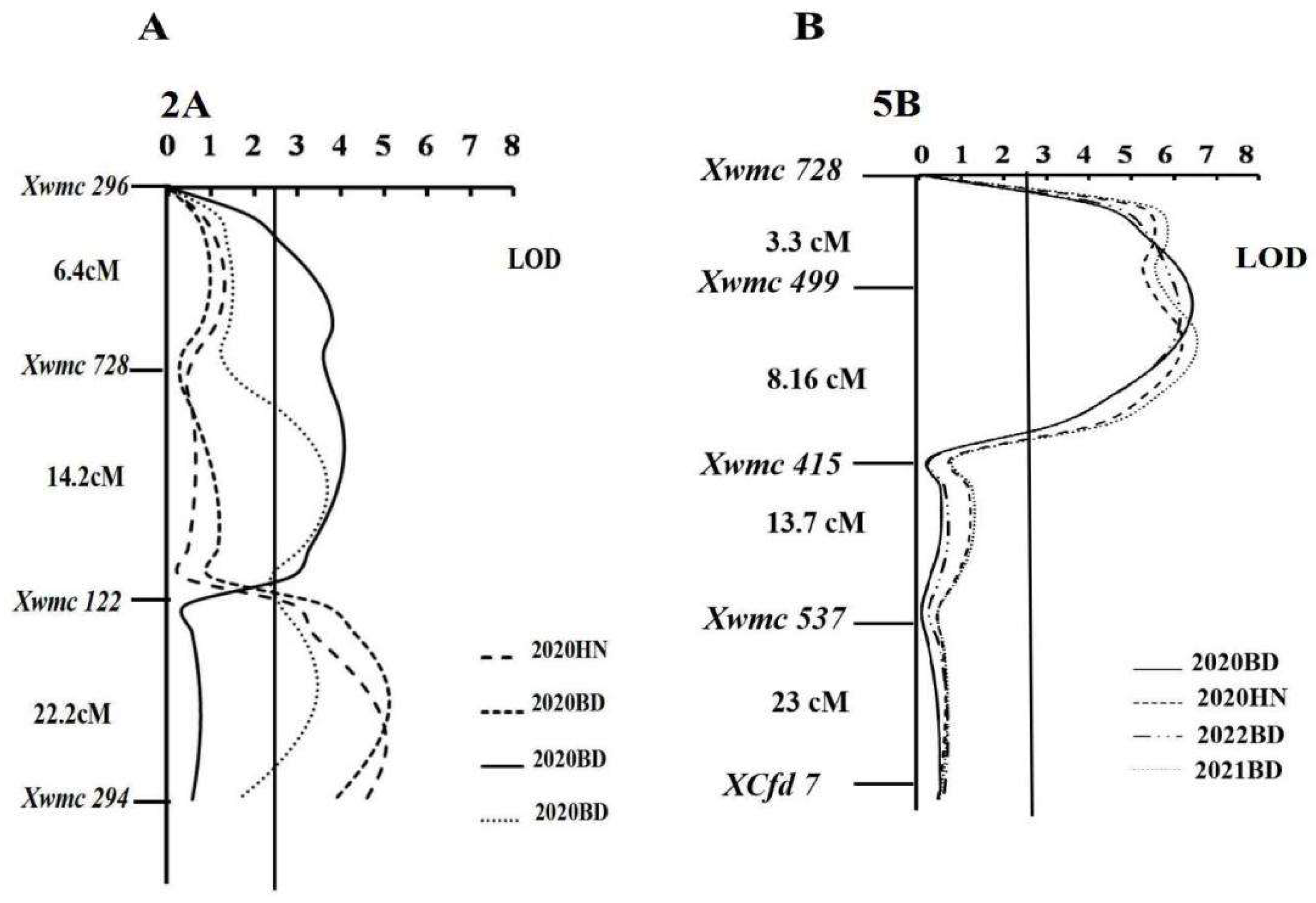
Lr.hbau-2AL.1, located between molecular markers
xgwm122-xwmc294 was identified in trials 2021BD, 2020BD, and 2020HN, explaining 14.4%, 12.2%, and 17.0%, of the phenotypic variance, respectively, in these trials (
Table 3; Figure 4).
QLr.hbau-2AL.2 was detected in the trials 2021BD and 2022BD between SSR markers
xwmc728-xgwm122 (Figure 4).
QLr.hbau-2A.2 explained 13.2% and 12.5% of the phenotypic variance in trials 2021BD and 2022BD, respectively, corresponding to minor effects (
Table 3).
QLr.hbau-5BL was an important QTL for leaf rust resistance in all environments, consistently explaining 17-19% of the phenotypic variances across all environments (
Table 3).
QLr.hbau-5BL was located between SSR markers
xgwm499-xwmc415. The results show that the alleles associated with leaf rust resistance within the QTLs on 2AL and 5AL were inherited from N. Strampelli.(Figure 4)
Figure 3.
Genetic maps of chromosome 2A(A) and 5B(B) of N. Strampelli×Huixianhong RIL population, illustrating the logarithm of the odds (LOD). Marker names and intervals (in Kosambi cM) between adjacent markers are shown along the chromosome orientated with the telomere of 2AL and the centromere of 5BL to the left. Horizontal lines indicate the threshold LOD of 2.5. Each of the small triangles along the x-axis represents a marker used for QTL mapping.
Figure 3.
Genetic maps of chromosome 2A(A) and 5B(B) of N. Strampelli×Huixianhong RIL population, illustrating the logarithm of the odds (LOD). Marker names and intervals (in Kosambi cM) between adjacent markers are shown along the chromosome orientated with the telomere of 2AL and the centromere of 5BL to the left. Horizontal lines indicate the threshold LOD of 2.5. Each of the small triangles along the x-axis represents a marker used for QTL mapping.
Table 3.
Puccinia triticina races identified from the field trial plots of Baoding in 2020.
Table 3.
Puccinia triticina races identified from the field trial plots of Baoding in 2020.
| Race |
Virulence (ineffective gene)a |
| THTT |
1, 2a, 2c, 3, 16, 26 ,3ka, 11, 17, 30, B, 10, 14a, 18 |
| PHPS |
1, 2c, 3, 16, 26, 3ka ,17, 30, B, 10, 14a |
| THTQ |
1, 2a, 2c, 3, 16, 26, 3ka, 11, 17, 30, B, 10 |
| THTS |
1, 2a, 2c, 3, 16, 26, 3ka, 11, 17, 30, B, 10, 14a, 3 |
Table 4.
APR QTLs associated with leaf rust in wheat N.Strampelli×Huixianhong RIL populations.
Table 4.
APR QTLs associated with leaf rust in wheat N.Strampelli×Huixianhong RIL populations.
| QTLa |
Environmentb |
Interval |
Positionc |
LODd |
PVE (%)e |
Addf |
| QLr.hbau-2AL.1 |
*2020BD |
Xgwm122-Xwmc294 |
19 |
4.1 |
12.2 |
−9.4 |
| QLr.hbau-5BL |
|
Xgwm499-Xwmc415 |
4 |
6.4 |
18.7 |
−10.6 |
| QLr.hbau-2AL.1 |
*2020 HN |
Xgwm122-Xwmc294 |
20 |
5.1 |
17 |
−9.3 |
| QLr.hbau-5BL |
|
Xgwm499-Xwmc415 |
5 |
6.5 |
19.2 |
−9.3 |
| QLr.hbau-2AL.1 |
*2021 BD |
Xgwm122-Xwmc294 |
18 |
3.5 |
14.4 |
−7.2 |
| QLr.hbau-2AL.2 |
|
Xwmc728-Xgwm122 |
11 |
3.7 |
13.2 |
−6.1 |
| QLr.hbau-5BL |
|
Xgwm499-Xwmc415 |
5 |
6.2 |
18.2 |
−9 |
| QLr.hbau-2AL.2 |
*2022 BD |
Xwmc728-Xgwm122 |
9 |
4.1 |
12.5 |
−7.9 |
| QLr.hbau-5BL |
|
Xgwm499-Xwmc415 |
4 |
6.1 |
17.9 |
−10.4 |
4. Discussion
Comparisons with previous reports of LR QTLs
QLr.hbau-2AL.1
To date, three APR QTLs
QLr.cimmyt-2AL,
QLr.
sfr-2AL, and
QLr.
ubo-2AL[
15,
16] were reported on chromosome 2A in a durum population.
QLr.cimmyt-2AL was located between SSR markers
wpt4419-wpt8226 in the Mexican wheat cultivar Avocet and explained 5.8-7.2% of the phenotypic variation[
17].
QLr.sfr-2AL was located between SSR markers
cfa2263c-sfr.be590525 in the Swiss bread-wheat cultivar Forno and explained 9.5-12% of the phenotypic variation[
18].
QLr.ubo-2AL was located between SSR markers
wpt-386-310911 in the North American durum-wheat cultivar Lloyd and explained 18.6-30% of the phenotypic variation[
19].
QLr.cimmyt-2AL and
QLr.sfr-2AL are located at 63 cM on chromosome 2A, while
QLr.ubo-2AL is located at 143 cM at the end of chromosome 2AL[
20]. Herein,
QLr.hbau-2AL.1 was detected in all the environments and explained 12-17% of the phenotypic variance, and the SSR markers area
xgwm122-xwmc294 is 20cM near the 2A centromere, and different from the aforemention.ed QTLs, suggesting that it might be a new QTL.
QLr.hbau-2AL.2
QLr.hbau-2AL.2 was detected in two the environments (2021BD and2022BD) and explained 12.5-13.2% of the phenotypic variance. To date, four APR genes,
Lr17,
Lr37, Lr45, and
Lr65 were mapped on chromosome 2A.
Lr37 was located at 10 cM near the centromere of 2AS chromosome[
21,
22]. Herein,
QLr.hbau-2AL.2 was located at 9 cM near the centromere of the 2AL chromosome of cultivar N. Strampelli. Similarly,
Lr37 was located at 10 cM near the centromere of 2AS chromosome. However,
Lr37 is a major gene, explaining high levels of variation, while
QLr.tbau-2AL.2 explained less than 15% of the variance in FDS. Therefore,
QLr.tbau-2AL.2 is unlikely to correspond to
Lr37 and may represent a novel site associated with leaf rust resistance and might be new.
QLr.hbau-5BL
QLr.hbau-5BL which was located on chromosome 5B between SSR markers
Xgwm499-Xwmc415 (genetic distance 8.1 cM; 477-507 Mb in the Chinese Spring reference mapbased on the position of the reference sequence) ,was strongly associated with leaf rust resistance. Four QTLs (
QLr.ccs-5b.4, QLr.ccs-5b.5, QLr.inra-5B, and
Lr.locus-5B.1) and two genes (
Lr18 and
Lr52) for LR two genes resistance have been mapped to chromosome 5B[
23,
24]. e..
QLr.Ccsu-5b.4 and
QLr.Ccsu-5b.5 were obtained using an Inclusive Composite Interval Mapping (ITMI) mapping population, with the SSR markers intervals
Xcdo1326-Xbarc140and
Xbarc142-Xbarc69[
25,
26].
QLr.Locus-5B.1 was identified between SSR markers
Xgwm433-Xgwm234 in the durum wheat variety Wollaroi which is resistant to leaf rust throughout the growth period, while
QLr.inra-5B obtained from double haploid (DH) population hybridized by Apache/balance varieties, was between the SSR markers
Wmc517-Wpt7720[
25]. Hence, the identified Lr.hbau-5BL with different marker intervals and from different genetic groups, is different from the aforementioned, and might be new.
Potentially pleiotropic QTLs associated with resistance to multiple wheat diseases
detected a QTL locus (QPM) of adult plant resistance to powdery mildew on 5BL in F2:3 generation of N. Strampelli×Huixianhong[
27]. The SSR markers interval was located in xbarc331-xwmc537 (608-953 Mb on the physical map). located the QTL locus on 5BL for the F2:3 population of N. Strampelli/Mingxian 169. The SSR marker
xgwm499-xwmc415 (477-507 Mb on the physical map) was similar to the stripe rust resistance gene
YrN.S and is closely linked. In this study,
QLr.hbau-5B was located between SSR markers
xgwm499-xwmc415 (477-507 Mb on the physical map). Thus, QTLs associated with resistance located on chromosome 5BL near
QLr.hbau-5BL may be a pleiotropic site with multiple effects.
Interactions among QTLs for leaf rust resistance in adult plants
Although several studies have reported major genes associated with disease resistance, few studies to date have focused on the effects of minor resistance genes because the many interactions among minor genes affect accurate QTL localization. However, it has been suggested that the effects produced by the aggregation of multiple minor-effect genes will be long lasting than major genes. The selection pressure exerted by minor-effect resistance genes on
Pt is low, and the pathogen is less likely to evolve defenses or be replaced by competitors. Thus, minor-effect resistance genes are more industrially valuable than major genes. In this study ,the RIL population of N. Strampelli×Huixianhong showed good horizontal resistance in the field, and the QTLs on 5BL are likely to be closely linked with the stripe-rust resistance gene
YrN.S. Thus, disease resistance in the RIL population is likely to be controlled by one major gene or several minor genes, but further study is needed to explore these two alternatives[
28].
The results of this study obtained using high density SNP markers are improvements over previous studies that were based only on SSR markers. These improvements might be because we used a wheat 660K SNP chip, which has many polymorphic markers for genotyping. Previous studies used only limited SSR markers and few chromosomes for QTL mapping and analysis.
Implications of QTLs detected in the RIL population
The Italian wheat variety N. Strampelli has shown high levels of resistance to leaf rust, stripe rust, and powdery mildew for many years. In this study, three QTLs for leaf rust, including three potentially pleiotropic QLr loci and closely linked SNP markers were detected in the N. Strampelli×Huixianhong RIL population. Among these, the novel adult plant resistanceQLr.hbau-5BL on chromosome 5B conferred stable resistance to leaf rust. QLr.hbau-5BL was also associated with resistance to leaf rust and stripe rust, and this QTL was closely linked to the stripe rust resistance gene YrN.S, although this association requires further verification. In future studies, the SNP markers linked with the two QTLs will be genotyped using KASP assays to facilitate the pyramiding of rust resistance loci into future cultivars, as well as marker-assisted selection (MAS) in wheat breeding programs.
CRediT authorship contribution statement
YL phenotyped the population in thefield, prepared genotypic data, analyzed the data, and wrote the initial manuscript. XL design this experiment.
Author Contributions
“Conceptualization, LI.Man. ; methodology, LI.Man.; software, LI.Xue.; validation, LI.Man.; formal analysis, GAO.Teng.; investigation, LI.Xue and LI.Man.; resources, LI.Xing.; data curation,ZHANG Jia-qi and LI.Man.; writing—original draft preparation, LI.Man.; writing—review and editing, LI.Man.;visualization, LI.Xue.; supervision, KANG Zhan-hai and LI.Xing ; project administration,KANG Zhan-hai;funding acquisition, LI.Xing. All authors have read and agreed to the published version of the manuscript.” Please turn to the CRediT taxonomy for the term explanation. Authorship must be limited to those who have contributed substantially to the work reported.
Funding
This research was funded by the National Natural Science Foundation of China (32001890), 2023 Scholarship Program for Introducing Overseas Students (C20230107).
Data Availability Statement
The original contributions presented in this study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author(s).
Acknowledgments
This study was sponsored by the National Natural Science Foundation of China (32001890), 2023 Scholarship Program for Introducing Overseas Students (C20230107).
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appearedn to influence the work reported in this paper.
Abbreviations
The following abbreviations are used in this manuscript:
| 2AL |
The long arm of chromosome 2A |
| 5BL |
The long arm of chromosome 5B |
| ANOVA |
Analysis of variance |
| APR |
Adult plant resistance |
| IT |
Infection type |
| LOD |
Logarithm of odds |
| Lr gene |
Leaf rust resistance gene |
| MAS |
Marker-assisted selection |
| PVE |
Phenotypic variation explained |
| QTL |
Quantitative trait locus/loci |
| RIL |
Recombinant inbred line |
| SNP |
Single nucleotide polymorphism |
| ICIM |
Inclusive Composite Interval Mapping |
| MDS |
The maximum disease severity |
References
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Bukhari, A.; Dar, Z.; Rizvi, S. Status and strategies in breeding for rust resistance in wheat. Agric. Sci. 2013, 4, 292–301. [Google Scholar] [CrossRef]
- Zhou, H.; Xia, X.; He, Z.; Li, X.; Wang, C.; Li, Z.; Liu, D. Molecular mapping of leaf rust resistance gene LrNJ97 in Chinese wheat line Neijiang 977671. Theor. Appl. Genet. 2013, 126, 2141–2147. [Google Scholar] [CrossRef]
- Prasad, P.; Savadi, S.; Bhardwaj, S.C.; Gupta, P.K. The progress of leaf rust research in wheat. Fungal Biol. 2020, 124, 537–550. [Google Scholar] [CrossRef]
- Lowe, I.; Jankuloski, L.; Chao, S.; Chen, X.; See, D.; Dubcovsky, J. Mapping and validation of QTL which confer partial resistance to broadly virulent post-2000 North American races of stripe rust in hexaploid wheat. Theor Appl Genet 2011, 123, 143–157. [Google Scholar] [CrossRef]
- Kumar, D.; Kumar, A.; Chhokar, V.; Gangwar, O.P.; Bhardwaj, S.C.; Sivasamy, M.; Prasad, S.V.S.; Prakasha, T.L.; Khan, H.; Singh, R.; Sharma, P.; Sheoran, S.; Iquebal, M.A.; Jaiswal, S.; Angadi, U.B.; Singh, G.; Rai, A.; Singh, G.P.; Kumar, D.; Tiwari, R. Genome-wide association studies in diverse spring wheat panel for stripe, stem, and leaf rust resistance. Front. Plant Sci. 2020, 11, 748. [Google Scholar] [CrossRef] [PubMed]
- Kolmer, J.A.; Bajgain, P.; Rouse, M.N.; Li, J.; Zhang, P. Mapping and characterization of the recessive leaf rust resistance gene Lr83 on wheat chromosome arm 1DS. Theor Appl Genet 2023, 136, 115. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Foessel, S.A.; Lagudah, E.S.; Huerta-Espino, J.; Hayden, M.J.; Bariana, H.S.; Singh, D.; Singh, R.P. New slow-rusting leaf rust and stripe rust resistance genes Lr67 and Yr46 in wheat are pleiotropic or closely linked. Theor Appl Genet, 2011, 122, 239–249. [Google Scholar] [CrossRef]
- Lan, C.X.; Singh, R.P.; Huerta-Espino, J.; Calvo-Salazar, V.; Herrera-Foessel, S.A. Genetic analysis of resistance to leaf rust and stripe rust in wheat cultivar Francolin#1. Plant Dis 2014, 98, 1227–1234. [Google Scholar] [CrossRef]
- Pinto da Silva, G.B.; Zanella, C.M.; Martinelli, J.A.; Chaves, M.S.; Hiebert, C.W.; McCallum, B.D.; Boyd, L.A. Quantitative trait loci conferring leaf rust resistance in hexaploid wheat. Phytopathology 2018, 108, 1344–1354. [Google Scholar] [CrossRef]
- Lu, Y.; Lan, C.; Liang, S.; Zhou, X.; Liu, D.; Zhou, G.; Lu, Q.; Jing, J.; Wang, M.; Xia, X.; He, Z. QTL mapping for adult-plant resistance to stripe rust in Italian common wheat cultivars Libellula and Strampelli. Theor Appl Genet 2009, 119, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.J.; Kreis, M.; Shewry, P.R.; Gale, M.D. Location of β-amylase sequences in wheat and its relatives. Theor. Appl. Genet. 1988, 75, 286–290. [Google Scholar] [CrossRef]
- Lan, C.; Liang, S.; Zhou, X.; Zhou, G.; Lu, Q.; Xia, X.; He, Z. Identification of genomic regions controlling adult-plant stripe rust resistance in Chinese landrace Pingyuan 50 through bulked segregant analysis. Phytopathology 2010, 100, 313–318. [Google Scholar] [CrossRef]
- Asad, M.A.; Bai, B.; Lan, C.; Yan, J.; Xia, X.; Zhang, Y.; He, Z. Identification of QTL for adult-plant resistance to powdery mildew in Chinese wheat landrace Pingyuan 50. Crop J. 2014, 2, 308–314. [Google Scholar] [CrossRef]
- Schnurbusch, T.; Paillard, S.; Schori, A.; Messmer, M.; Schachermayr, G.; Winzeler, M.; Keller, B. Dissection of quantitative and durable leaf rust resistance in Swiss winter wheat reveals a major resistance QTL in the Lr34 chromosomal region. Theor Appl Genet. 2004, 108, 477–484. [Google Scholar] [CrossRef]
- Rosewarne, G.M.; Singh, R.P.; Huerta-Espino, J.; Rebetzke, G.J. Quantitative trait loci for slow-rusting resistance in wheat to leaf rust and stripe rust identified with multi-environment analysis. Theor Appl Genet 2008, 116, 1027–1034. [Google Scholar] [CrossRef]
- Rosewarne, G.M.; Singh, R.P.; Huerta-Espino, J.; Herrera-Foessel, S.A.; Forrest, K.L.; Hayden, M.J.; Rebetzke, G.J. Analysis of leaf and stripe rust severities reveals pathotype changes and multiple minor QTLs associated with resistance in an Avocet × Pastor wheat population. Theor Appl Genet 2012, 124, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Schnurbusch, T.; Paillard, S.; Schori, A.; Messmer, M.; Schachermayr, G.; Winzeler, M.; Keller, B. Dissection of quantitative and durable leaf rust resistance in Swiss winter wheat reveals a major resistance QTL in the Lr34 chromosomal region. Theor Appl Genet 2004, 108, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Mantovani, P.; Tuberosa, R.; Deambrogio, E.; Giuliani, S.; Demontis, A.; Massi, A.; Sanguineti, M.C. A major QTL for durable leaf rust resistance widely exploited in durum wheat breeding programs maps on the distal region of chromosome arm 7BL. Theor Appl Genet. 2008, 117, 1225–1240. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lan, C.; He, Z.; Singh, R.P.; Rosewarne, G.M.; Chen, X.; Xia, X. Overview and Application of QTL for Adult Plant Resistance to Leaf Rust and Powdery Mildew in Wheat. Crop Science 2014, 54, 1907. [Google Scholar] [CrossRef]
- Kloppers, F.J.; Pretorius, Z.A. Effects of combinations amongst genes Lr13, Lr34 and Lr37 on components of resistance in wheat to leaf rust. Plant Pathol. 1997, 46, 737–750. [Google Scholar] [CrossRef]
- Mohler, V.; Singh, D.; Singrün, C.; Park, R.F. Characterization and mapping of Lr65 in spelt wheat ‘Altgold Rotkorn. Plant Breed. 2012, 131, 252–257. [Google Scholar] [CrossRef]
- Yamamori, M. An N-band marker for gene Lr18 for resistance to leaf rust in wheat. Theor Appl Genet 1994, 89, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Juliana, P.; Singh, R.P.; Singh, P.K.; Poland, J.A.; Bergstrom, G.C.; Huerta-Espino, J.; Bhavani, S.; Crossa, J.; Sorrells, M.E. Genome-wide association mapping for resistance to leaf rust, stripe rust and tan spot in wheat reveals potential candidate genes. Theor. Appl. Genet. 2018, 131, 1405–1422. [Google Scholar] [CrossRef]
- Azzimonti, G.; Marcel, T.C.; Robert, O.; Paillard, S.; Lannou, C.; Goyeau, H. Diversity, specificity and impacts on field epidemics of QTLs involved in components of quantitative resistance in the wheat leaf rust pathosystem. Mol. Breed. 2014, 34, 549–567. [Google Scholar] [CrossRef]
- Aoun, M.; Breiland, M.; Kathryn Turner, M.; Loladze, A.; Chao, S.; Xu, S.S.; Ammar, K.; Anderson, J.A.; Kolmer, J.A.; Acevedo, M. Genome-wide association mapping of leaf rust response in a durum wheat worldwide germplasm collection. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Azeem, A.M.; Bai, B.; Lan, C.; Yan, J.; Xia, C.; Zhang, Y.; He, Z. QTL Mapping for adult plant resistance to powdery mildew in Italian wheat cv. Strampelli. J. Integr. Agric. 2013, 5, 756–764. [Google Scholar] [CrossRef]
- William, H.M.; Singh, R.P.; Huerta-Espino, J.; Palacios, G.; Suenaga, K. Characterization of genetic loci conferring adult plant resistance to leaf rust and stripe rust in spring wheat. Genome 2006, 49, 977–990. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).