1. Introduction
CRISPR-Cas systems have emerged as transformative tools in genetic engineering, with impactful applications in the treatment of several challenging human diseases. This review focuses on five such diseases—Duchenne muscular dystrophy, Alzheimer's disease, HIV, cystic fibrosis, and sickle cell disease—chosen for their clinical importance and the diversity they offer in terms of gene-editing targets and therapeutic strategies. By analyzing these cases, we aim to provide insight into how CRISPR technologies are shaping modern approaches to precision medicine. Initially discovered as an adaptive immune system in bacteria, CRISPR systems have now been adapted for diverse biomedical applications, from disease modeling to therapeutic interventions. While Cas9 remains the most commonly used, recent studies have shown growing interest in Cas12 for its high-fidelity DNA editing, Cas13 for RNA-targeting potential, and the newly emerging Cas15 for its compact structure and programmable RNA cleavage. This review aims to contextualize the relevance of these diseases and CRISPR tools in the broader scope of gene therapy advancements.
2. Methodology
We conducted a targeted literature search on PubMed covering the period from January 2015 to March 2025. Search terms included "CRISPR", "CRISPR-Cas9", "CRISPR-Cas12", "CRISPR-Cas13", "CRISPR-Cas15", along with each disease name (e.g., "Duchenne muscular dystrophy CRISPR"). We manually screened the search results to identify articles that focused on experimental therapeutic applications of CRISPR technologies. Inclusion criteria were: relevance to human disease therapy, clarity of methodology, and peer-reviewed status. From the shortlisted articles, we extracted information on disease targets, gene editing strategy, CRISPR variant used, experimental system (e.g., in vitro, animal model, clinical), and outcomes. Data was visualized to illustrate usage frequency and CRISPR system distribution.
3. Summary of CRISPR Applications in Selected Diseases
| Disease |
Target Gene |
CRISPR Type(s) |
System Used |
Outcome Summary |
| Duchenne Muscular Dystrophy |
DMD |
Cas9, Cas12, Prime Editing |
Mouse model |
Exon skipping, partial functional recovery |
| HIV |
CCR5 |
Cas9, Cas13 |
Humanized mice |
Resistance to HIV replication, potential transcriptome editing |
| Alzheimer's Disease |
BACE1 |
Cas9, Cas13, Cas15 |
Mouse model |
Reduced amyloid-beta accumulation; RNA-level modulation being explored |
| Cystic Fibrosis |
CFTR |
Cas9, Base Editing |
Organoid/in vitro |
Corrected mutation, restored protein function |
| Sickle Cell Disease |
HBB |
Cas9, Prime Editing |
Preclinical, stem cells |
Corrected sickling mutation with high precision |
These studies demonstrate both the therapeutic potential and methodological diversity of CRISPR-based interventions. While all studies showed promising outcomes, they differed in experimental setup and editing goals. For instance, Duchenne muscular dystrophy models focused on exon skipping for partial functional recovery, whereas cystic fibrosis studies aimed at precise mutation correction using organoids. HIV research prioritized conferring viral resistance through CCR5 editing, while Alzheimer's studies focused on reducing amyloid-beta levels. Such differences underscore the versatility of CRISPR tools in targeting diverse genetic mechanisms across diseases.
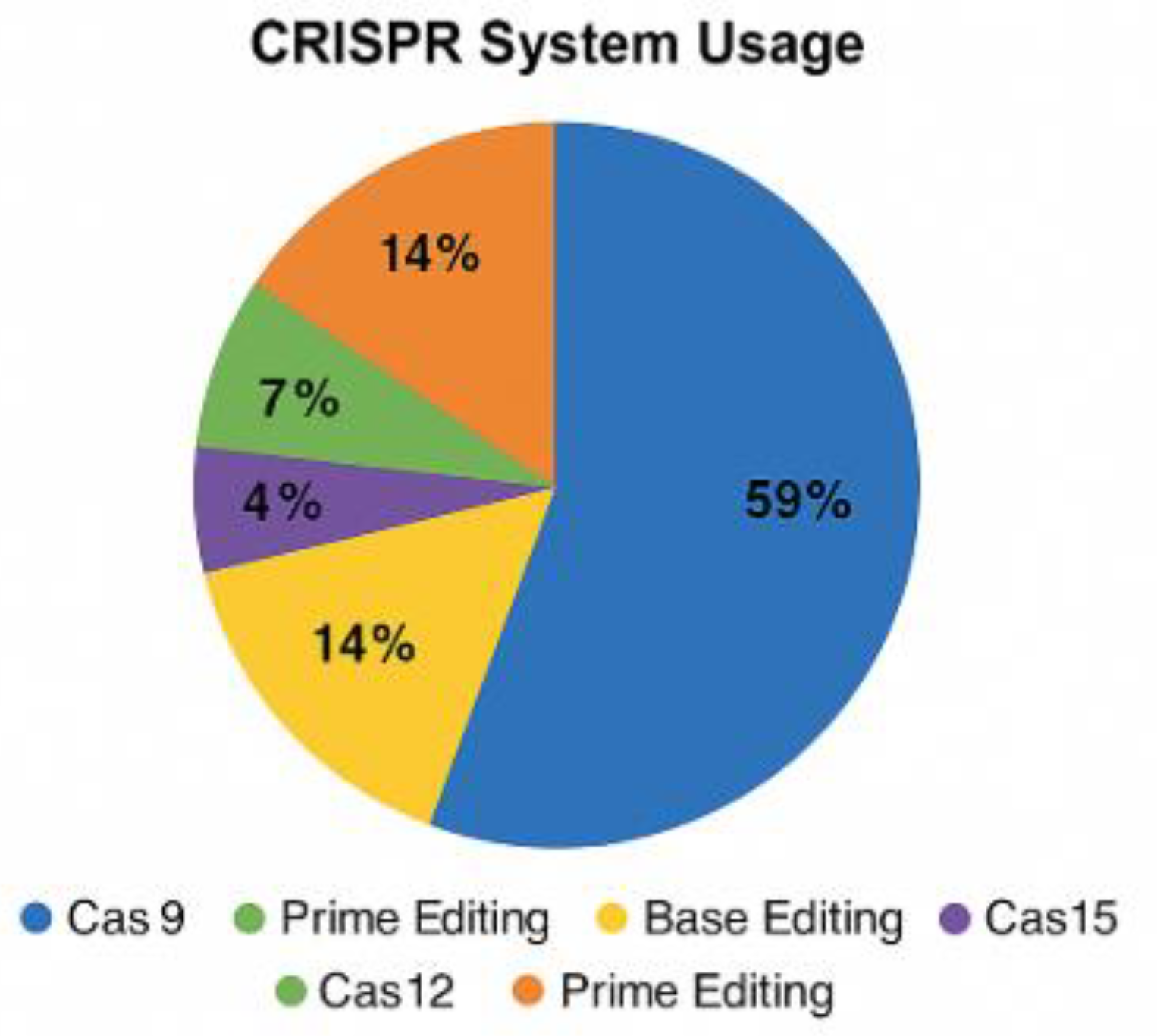
4. CRISPR Systems Usage Chart
Below is a visual representation of CRISPR usage frequency across the five studied diseases:
5. Discussion
Cas9 remains the most frequently used CRISPR system in therapeutic research due to its established protocols and widespread availability. However, Cas12 has gained attention for its compact size, staggered DNA cleavage patterns, and better specificity, making it a promising candidate for gene therapies requiring higher fidelity. Cas13 stands out for RNA editing applications—particularly useful in diseases where reversible gene expression control is advantageous. For example, Cas13 has been successfully applied in neurological disorders and viral diseases, where transcriptome-level interventions offer safety benefits.
Cas15, a newly discovered RNA-guided nuclease, is notable for its extremely small size and RNA-targeting precision. While studies are still emerging, its potential for delivery in compact viral vectors and synthetic systems makes it an exciting candidate for future gene therapy platforms. For instance, its programmable RNA cleavage has shown early promise in models of viral infection and in modulating gene expression in hard-to-target tissues.
Together, these systems open new possibilities for tailored editing based on disease pathology, molecular target, and desired outcomes. Base and prime editing technologies further enhance specificity and safety by enabling precise nucleotide conversions without double-stranded breaks. Ongoing research into improved delivery systems—viral and non-viral—remains essential for clinical translation.
6. Limitations
This review captures only a fraction of the rapidly evolving CRISPR research landscape, focusing on five specific diseases to illustrate current trends. Many significant studies were excluded due to space and scope limitations. A comprehensive meta-analysis would be needed to derive generalized conclusions about CRISPR efficacy, safety, and comparative performance of different Cas systems. Furthermore, some systems like Cas15 are still in their infancy, and their clinical potential remains to be fully validated. Future reviews should expand to include quantitative data synthesis, broader disease models, and integration of multi-omic approaches to evaluate therapeutic potential. Future directions should also prioritize long-term safety studies, robust delivery methods, and the combination of CRISPR with AI-driven predictive models to enhance precision.
7. Conclusion
CRISPR technology is revolutionizing molecular medicine, offering new hope for the treatment of complex human diseases. Even within this focused review, the versatility and therapeutic potential of CRISPR-based tools are clear—ranging from correcting single-gene disorders like sickle cell disease to targeting complex conditions such as Alzheimer's and HIV.
While Cas9 continues to dominate current applications, the rise of Cas12, Cas13, and Cas15—alongside base editing and prime editing techniques—signals a new wave of innovation. These next-generation tools not only improve editing precision but also open the door to non-permanent and highly specific interventions, especially when paired with advances in delivery systems such as lipid nanoparticles and viral vectors.
Looking ahead, CRISPR’s integration into clinical practice will depend on overcoming delivery challenges, minimizing off-target effects, and ensuring long-term safety. Continued interdisciplinary collaboration between geneticists, bioengineers, and clinicians is essential.
Future research should prioritize large-scale clinical trials, refined delivery vehicles, and deeper exploration of CRISPR’s RNA-targeting and epigenetic modulation capacities. With sustained innovation and responsible deployment, CRISPR technologies hold the promise to usher in a new era of precision medicine tailored to individual patient needs.
Author Contributions
P. Pooja Shree performed data curation and literature review. Paper independently compiled and drafted by author.
Acknowledgements
We thank the open-source community and the original authors of each reviewed paper.
Conflicts of Interest
None declared.
References
- Long C, et al. "Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy.". Science, 2016, 351, 400–403. [CrossRef] [PubMed]
- Kang H, et al. "CRISPR-Cas9 mediates gene editing of CCR5 in human primary T cells for HIV resistance.". Molecular Therapy, 2017, 25, 1782–1792. [CrossRef] [PubMed]
- Park H, et al. "CRISPR-Cas9 editing of BACE1 reduces beta-amyloid in Alzheimer’s disease mouse models.". Nature Neuroscience, 2019, 22, 547–556.
- Schwank G, et al. "Functional repair of CFTR by CRISPR-Cas9 in intestinal stem cell organoids from cystic fibrosis patients.". Cell Stem Cell, 2013, 13, 653–658. [CrossRef] [PubMed]
- Dever DP, et al. "CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells.". Nature, 2016, 539, 384–389. [CrossRef] [PubMed]
- Frangoul H, et al. "CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia.". New England Journal of Medicine, 2021, 384, 252–260. [CrossRef] [PubMed]
- Chen JS, et al. "CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity.". Science, 2018, 360, 436–439. [CrossRef] [PubMed]
- Abudayyeh OO, et al. "C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector.". Science, 2016, 353, aaf5573. [CrossRef] [PubMed]
- Cox DBT, et al. “RNA editing with CRISPR-Cas13.”. Science, 2017, 358, 1019–1027. [CrossRef] [PubMed]
- Gaudelli NM, et al. “Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage.”. Nature, 2017, 551, 464–471. [CrossRef] [PubMed]
- Anzalone AV, et al. “Search-and-replace genome editing without double-strand breaks or donor DNA.”. Nature, 2019, 576, 149–157. [CrossRef] [PubMed]
- Yan WX, et al. “Cas15 is a compact and efficient RNA-guided RNA nuclease.”. Nature Biotechnology, 2022, 40, 250–258.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).