Submitted:
15 April 2025
Posted:
15 April 2025
You are already at the latest version
Abstract
Keywords:
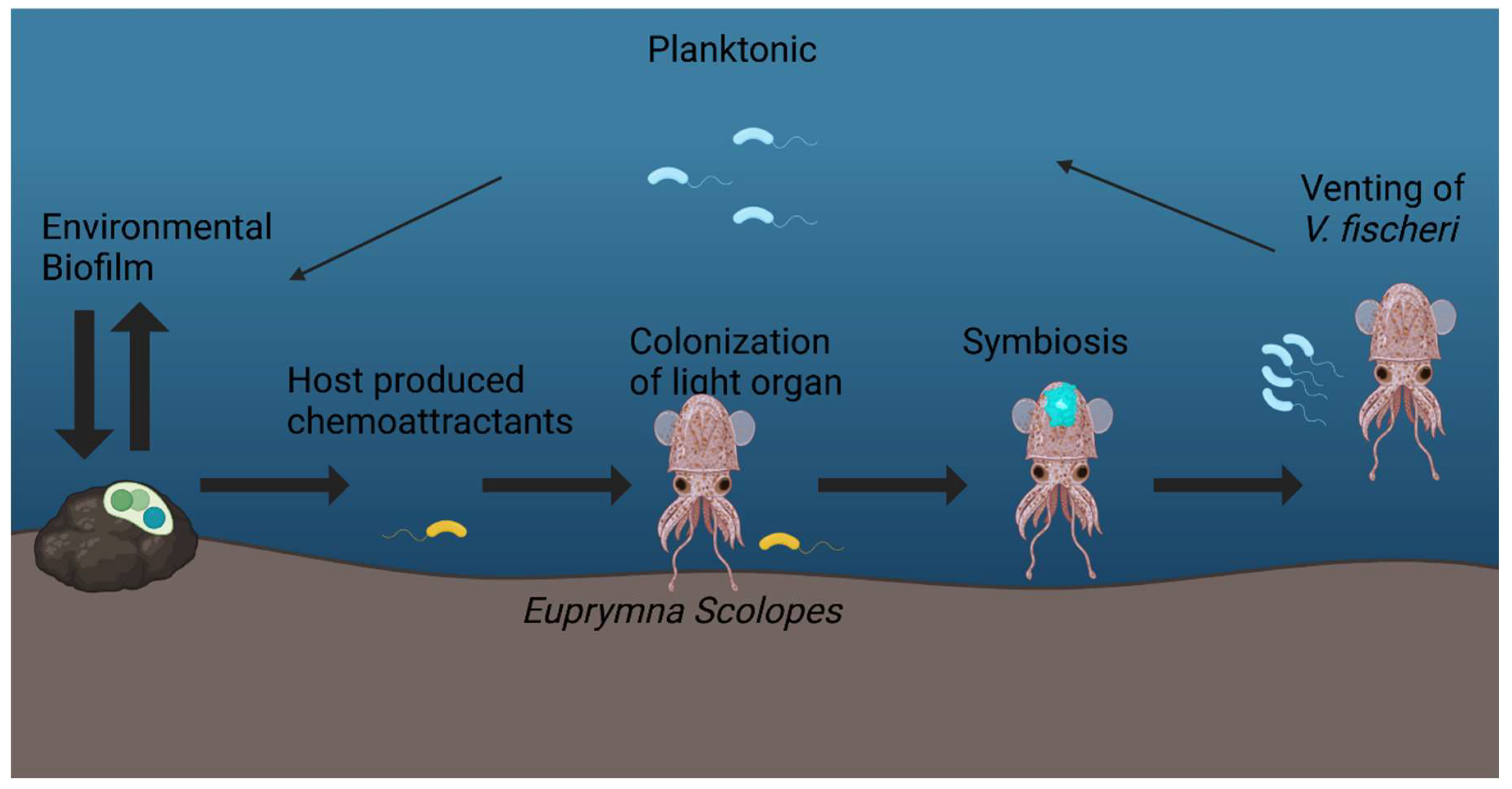
1. Introduction
2. Biofilms: Essential Structures in Symbiosis
3. Vibrio Fischeri as a Model for the Study of Host Associated Biofilms
4. Expanding the Lens: Other Vibrio and Photobacterium Species
5. Adaptive and Functional Roles of Biofilms in Symbiosis
6. Advances in Understanding Marine Biofilms
7. Conclusion and Future Directions
References
- Nyholm, S. V; McFall-Ngai, M.J. A lasting symbiosis: how the Hawaiian bobtail squid finds and keeps its bioluminescent bacterial partner. Nat Rev Microbiol 2021, 19, 666–679. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.W.; Nishiguchi, M.K. Counterillumination in the hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar Biol 2004, 144, 1151–1155. [Google Scholar] [CrossRef]
- Fung, B.L.; Esin, J.J.; Visick, K.L. Vibrio fischeri : a model for host-associated biofilm formation. J Bacteriol 2024, 206. [Google Scholar] [CrossRef] [PubMed]
- Nyholm, S. V.; McFall-Ngai, M. The winnowing: establishing the squid–Vibrio symbiosis. Nat Rev Microbiol 2004, 2, 632–642. [Google Scholar] [CrossRef]
- Soto, W.; Nishiguchi, M.K. Microbial experimental evolution as a novel research approach in the Vibrionaceae and squid-Vibrio symbiosis. Front Microbiol 2014, 5. [Google Scholar] [CrossRef]
- Soto, W.; Nishiguchi, M.K. Environmental stress selects for innovations that drive Vibrio symbiont diversity. Front Ecol Evol 2021, 9. [Google Scholar] [CrossRef]
- Chavez-Dozal, A.; Hogan, D.; Gorman, C.; Quintanal-Villalonga, A.; Nishiguchi, M.K. Multiple Vibrio fischeri genes are involved in biofilm formation and host colonization. FEMS Microbiol Ecol 2012, 81, 562–573. [Google Scholar] [CrossRef]
- Nourabadi, N.; Nishiguchi, M.K. Ph adaptation drives diverse phenotypes in a beneficial bacterium-host mutualism. Front Ecol Evol 2021, 9. [Google Scholar] [CrossRef]
- Chavez-Dozal, A.A.; Gorman, C.; Lostroh, C.P.; Nishiguchi, M.K. Gene-swapping mediates host specificity among symbiotic bacteria in a beneficial symbiosis. PLoS One 2014, 9, e101691. [Google Scholar] [CrossRef]
- Visick, K.L.; Schembri, M.A.; Yildiz, F.; Ghigo, J.-M. Biofilms 2015: multidisciplinary approaches shed light into microbial life on surfaces. J Bacteriol 2016, 198, 2553–2563. [Google Scholar] [CrossRef]
- Yildiz, F.H.; Visick, K.L. Vibrio biofilms: so much the same yet so different. Trends Microbiol 2009, 17, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Lovell, C.R. Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol Biol Rev 2016, 80, 91–138. [Google Scholar] [CrossRef]
- Takemura, A.F.; Chien, D.M.; Polz, M.F. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front Microbiol 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Zamborsky, D.J.; Nishiguchi, M.K. Phylogeographical patterns among mediterranean Sepiolid squids and their Vibrio symbionts: environment drives specificity among sympatric species. Appl Environ Microbiol 2011, 77, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Dozal, A.; Gorman, C.; Erken, M.; Steinberg, P.D.; McDougald, D.; Nishiguchi, M.K. Predation response of Vibrio fischeri biofilms to bacterivorus protists. Appl Environ Microbiol 2013, 79, 553–558. [Google Scholar] [CrossRef]
- Chavez-Dozal, A.; Gorman, C.; Nishiguchi, M.K. Proteomic and metabolomic profiles demonstrate variation among free-living and symbiotic Vibrio fischeri biofilms. BMC Microbiol 2015, 15, 226. [Google Scholar] [CrossRef]
- Chavez-Dozal, A.; Soto, W.; Nishiguchi, M.K. Identification of a transcriptomic network underlying the wrinkly and smooth phenotypes of Vibrio fischeri. J Bacteriol 2021, 203. [Google Scholar] [CrossRef]
- Yanovski, R.; Barak, H.; Brickner, I.; Kushmaro, A.; Abelson, A. The microbial community of coral reefs: biofilm composition on artificial substrates under different environmental conditions. Mar Biol 2024, 171, 74. [Google Scholar] [CrossRef]
- de Carvalho, C.C.C.R. Marine Biofilms: A successful microbial strategy with economic implications. Front Mar Sci 2018, 5. [Google Scholar] [CrossRef]
- Mao-Jones, J.; Ritchie, K.B.; Jones, L.E.; Ellner, S.P. How microbial community composition regulates coral disease development. PLoS Biol 2010, 8, e1000345. [Google Scholar] [CrossRef]
- Broszat, M.; Grohmann, E. Horizontal gene transfer in planktonic and biofilm modes. In; 2014; pp. 67–95.
- Madsen, J.S.; Burmølle, M.; Hansen, L.H.; Sørensen, S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol 2012, 65, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, M.; Brooks, B.; Brooks, A. The complex relationship between virulence and antibiotic resistance. Genes (Basel) 2017, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Ghazvinian, M.; Asgharzadeh Marghmalek, S.; Gholami, M.; Amir Gholami, S.; Amiri, E.; Goli, H.R. Antimicrobial resistance patterns, virulence genes, and biofilm formation in enterococci strains collected from different sources. BMC Infect Dis 2024, 24, 274. [Google Scholar] [CrossRef]
- Soto, W.; Gutierrez, J.; Remmenga, M.D.; Nishiguchi, M.K. Salinity and temperature effects on physiological responses of Vibrio fischeri from diverse ecological niches. Microb Ecol 2009, 57, 140–150. [Google Scholar] [CrossRef]
- Soto, W.; Rivera, F.M.; Nishiguchi, M.K. Ecological diversification of Vibrio fischeri serially passaged for 500 generations in novel squid host Euprymna tasmanica. Microb Ecol 2014, 67, 700–721. [Google Scholar] [CrossRef]
- McFall, A.; Coughlin, S.A.; Hardiman, G.; Megaw, J. Strategies for biofilm optimization of plastic-degrading microorganisms and isolating biofilm formers from plastic-contaminated environments. Sustain Microbiol 2024, 1. [Google Scholar] [CrossRef]
- Mitra, A.; Mukhopadhyay, S. Biofilm mediated decontamination of pollutants from the environment. AIMS Bioeng 2016, 3, 44–59. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, T.; Shaw, P.; Wang, P.-Y. Manipulating bacterial biofilms using materiobiology and synthetic biology approaches. Front Microbiol 2022, 13. [Google Scholar] [CrossRef]
- Mishra, A.; Aggarwal, A.; Khan, F. Medical device-associated infections caused by biofilm-forming microbial pathogens and controlling strategies. Antibiotics 2024, 13, 623. [Google Scholar] [CrossRef]
- Machineni, L. Effects of biotic and abiotic factors on biofilm growth dynamics and their heterogeneous response to antibiotic challenge. J Biosci 2020, 45, 25. [Google Scholar] [CrossRef]
- Chavez-Dozal, A.; Nishiguchi, M.K. Variation in biofilm formation among symbiotic and free-living strains of Vibrio fischeri. J Basic Microbiol 2011, 51, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Lu, W.; Rabinowitz, J.D.; Bassler, B.L. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of VpsT. J Bacteriol 2008, 190, 2527–2536. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Park, S.; Lee, K. Role of ntrc-regulated exopolysaccharides in the biofilm formation and pathogenic interaction of Vibrio vulnificus. Mol Microbiol 2009, 74, 436–453. [Google Scholar] [CrossRef] [PubMed]
- Urbanczyk, H.; Ast, J.C.; Dunlap, P. V. Phylogeny, genomics, and symbiosis of Photobacterium. FEMS Microbiol Rev 2011, 35, 324–342. [Google Scholar] [CrossRef]
- Battin, T.J.; Kaplan, L.A.; Denis Newbold, J.; Hansen, C.M.E. Contributions of microbial biofilms to ecosystem processes in stream mesocosms. Nature 2003, 426, 439–442. [Google Scholar] [CrossRef]
- Koch, E.J.; Moriano-Gutierrez, S.; Ruby, E.G.; McFall-Ngai, M.; Liebeke, M. The impact of persistent colonization by Vibrio fischeri on the metabolome of the host squid Euprymna scolopes. J Exp Biol 2020, 223. [Google Scholar] [CrossRef]
- Fung, B.L.; Visick, K.L. LitR and its quorum-sensing regulators modulate biofilm formation by Vibrio fischeri. J Bacteriol 2025. [Google Scholar] [CrossRef]
- Septer, A.N.; Visick, K.L. Lighting the way: how the Vibrio fischeri model microbe reveals the complexity of earth’s “simplest” life forms. J Bacteriol 2024, 206. [Google Scholar] [CrossRef]
- Pan, M.; Schwartzman, J.A.; Dunn, A.K.; Lu, Z.; Ruby, E.G. A single host-derived glycan impacts key regulatory nodes of symbiont metabolism in a coevolved mutualism. mBio 2015, 6. [Google Scholar] [CrossRef]
- Wang, Y.; Dufour, Y.S.; Carlson, H.K.; Donohue, T.J.; Marletta, M.A.; Ruby, E.G. H-nox–mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri. Proc Nat Acad Sci 2010, 107, 8375–8380. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.C.; Miyashiro, T. Quorum sensing in the squid-Vibrio symbiosis. Int J Mol Sci 2013, 14, 16386–16401. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Lade, H.; Paul, D.; Kweon, J.H. N-acyl homoserine lactone-mediated quorum sensing with special reference to use of quorum quenching bacteria in membrane biofouling control. Biomed Res Int 2014, 2014, 162584. [Google Scholar] [CrossRef]
- Shibata, S.; Yip, E.S.; Quirke, K.P.; Ondrey, J.M.; Visick, K.L. Roles of the structural symbiosis polysaccharide (syp) genes in host colonization, biofilm formation, and polysaccharide biosynthesis in Vibrio fischeri. J Bacteriol 2012, 194, 6736–6747. [Google Scholar] [CrossRef]
- Yip, E.S.; Geszvain, K.; DeLoney-Marino, C.R.; Visick, K.L. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol 2006, 62, 1586–1600. [Google Scholar] [CrossRef]
- Browne-Silva, J.; Nishiguchi, M.K. Gene sequences of the pil operon reveal relationships between symbiotic strains of Vibrio fischeri. Int J Syst Evol Microbiol 2008, 58, 1292–1299. [Google Scholar] [CrossRef]
- Darnell, C.L.; Hussa, E.A.; Visick, K.L. The putative hybrid sensor kinase sypf coordinates biofilm formation in Vibrio fischeri by acting upstream of two response regulators, SypG and VpsR. J Bacteriol 2008, 190, 4941–4950. [Google Scholar] [CrossRef]
- Millikan, D.S.; Ruby, E.G. Vibrio fischeri flagellin a is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J Bacteriol 2004, 186, 4315–4325. [Google Scholar] [CrossRef]
- Colin, R.; Ni, B.; Laganenka, L.; Sourjik, V. Multiple functions of flagellar motility and chemotaxis in bacterial physiology. FEMS Microbiol Rev 2021, 45. [Google Scholar] [CrossRef]
- Ariyakumar, D.S.; Nishiguchi, M.K. Characterization of two host-specific genes, mannose-sensitive hemagglutinin (MshA) and Uridyl phosphate dehydrogenase (UDPDH) that are involved in the Vibrio Fischeri-Euprymna tasmanica mutualism. FEMS Microbiol Lett 2009, 299, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Nishiguchi, M.K.; Hirsch, A.M.; Devinney, R.; Vedantam, G.; Riley, M.A.; Mansky, L.M. Deciphering evolutionary mechanisms between mutualistic and pathogenic symbioses. Vie et Milieu 2008, 58, 87–106. [Google Scholar] [PubMed]
- Bellissimo, K.A.; Septer, A.N.; Whistler, C.A.; Rodríguez, C.; Stabb, E. V. Deletion of luxI increases luminescence of Vibrio fischeri. mBio 2024, 15. [Google Scholar] [CrossRef] [PubMed]
- Plate, L.; Marletta, M.A. Nitric oxide modulates bacterial biofilm formation through a multicomponent cyclic-di-GMP signaling network. Mol Cell 2012, 46, 449–460. [Google Scholar] [CrossRef]
- Silva, A.J.; Benitez, J.A. Vibrio cholerae biofilms and cholera pathogenesis. PLoS Negl Trop Dis 2016, 10, e0004330. [Google Scholar] [CrossRef]
- He, H.; Cooper, J.N.; Mishra, A.; Raskin, D.M. Stringent response regulation of biofilm formation in Vibrio cholerae. J Bacteriol 2012, 194, 2962–2972. [Google Scholar] [CrossRef]
- Lutz, C.; Erken, M.; Noorian, P.; Sun, S.; McDougald, D. Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front Microbiol 2013, 4. [Google Scholar] [CrossRef]
- Conner, J.G.; Teschler, J.K.; Jones, C.J.; Yildiz, F.H. Staying alive: Vibrio cholerae ’s cycle of environmental survival, transmission, and dissemination. Microbiol Spectr 2016, 4. [Google Scholar] [CrossRef]
- Leighton, R.E.; Xiong, L.; Anderson, G.K.; Astarita, G.M.; Cai, G.; Norman, R.S.; Decho, A.W. Vibrio parahaemolyticus and Vibrio vulnificus in vitro biofilm dispersal from microplastics influenced by simulated human environment. Front Microbiol 2023, 14. [Google Scholar] [CrossRef]
- Chen, T.; Pu, M.; Subramanian, S.; Kearns, D.; Rowe-Magnus, D. PlzD Modifies Vibrio vulnificus foraging behavior and virulence in response to elevated c-di-GMP. mBio 2023, 14. [Google Scholar] [CrossRef]
- Lydon, K.A.; Kinsey, T.; Le, C.; Gulig, P.A.; Jones, J.L. Biochemical and virulence characterization of Vibrio vulnificus isolates from clinical and environmental sources. Front Cell Infect Microbiol 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Haygood, M.G.; Distel, D.L. Bioluminescent symbionts of flashlight fishes and deep-sea anglerfishes form unique lineages related to the genus Vibrio. Nature 1993, 363, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Tanet, L.; Tamburini, C.; Baumas, C.; Garel, M.; Simon, G.; Casalot, L. Bacterial bioluminescence: light emission in Photobacterium phosphoreum is not under quorum-sensing control. Front Microbiol 2019, 10. [Google Scholar] [CrossRef]
- Kitts, G.; Rogers, A.; Teschler, J.K.; Park, J.H.; Trebino, M.A.; Chaudry, I.; Erill, I.; Yildiz, F.H. The Rvv two-component regulatory system regulates biofilm formation and colonization in Vibrio cholerae. PLoS Pathog 2023, 19, e1011415. [Google Scholar] [CrossRef] [PubMed]
- Marsden, A.E.; Grudzinski, K.; Ondrey, J.M.; DeLoney-Marino, C.R.; Visick, K.L. Impact of salt and nutrient content on biofilm formation by Vibrio fischeri. PLoS One 2017, 12, e0169521. [Google Scholar] [CrossRef]
- Ramos-Vivas, J.; Acosta, F. Editorial: Host-bacteria interactions in fish pathogens. Front Cell Infect Microbiol 2024, 14. [Google Scholar] [CrossRef]
- Mass, S.; Cohen, H.; Podicheti, R.; Rusch, D.B.; Gerlic, M.; Ushijima, B.; van Kessel, J.C.; Bosis, E.; Salomon, D. The coral pathogen Vibrio coralliilyticus uses a T6SS to secrete a group of novel anti-eukaryotic effectors that contribute to virulence. PLoS Biol 2024, 22, e3002734. [Google Scholar] [CrossRef]
- Zhao, W.; Chen, X.; Liu, R.; Tian, P.; Niu, W.; Zhang, X.-H.; Liu, J.; Wang, X. Distinct coral environments shape the dynamic of planktonic Vibrio spp. Environ Microbiome 2023, 18, 77. [Google Scholar] [CrossRef]
- Mincer, T.J.; Zettler, E.R.; Amaral-Zettler, L.A. Biofilms on plastic debris and their influence on marine nutrient cycling, productivity, and hazardous chemical mobility. In Hazardous chemicals associated with plastics in the marine environment; Springer: Cham; vol 78, pp. 221–233.
- Zhang, X.; Lin, H.; Wang, X.; Austin, B. Significance of Vibrio species in the marine organic carbon cycle—a review. Sci China Earth Sci 2018, 61, 1357–1368. [Google Scholar] [CrossRef]
- Alotaibi, G.F. Factors influencing bacterial biofilm formation and development. Am J Biomed Sci Res 2021, 12, 617–626. [Google Scholar] [CrossRef]
- Filippini, G.; Bugnot, A.B.; Varkey, D.R.; Siboni, N.; Ferguson, A.; Gribben, P.E.; Erickson, K.; Palmer, J.; Dafforn, K.A. Nitrogen-cycling genes in oyster reefs and surrounding sediments: relationships with environmental factors and respective nitrogen rates. Mar Pollut Bull 2023, 197, 115710. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.E.; O’toole, G.A. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 2000, 64, 847–867. [Google Scholar] [CrossRef] [PubMed]
- McCaughey, C.S.; Trebino, M.A.; McAtamney, A.; Isenberg, R.Y.; Mandel, M.J.; Yildiz, F.H.; Sanchez, L.M. A Label-free approach for relative spatial quantitation of c-di-GMP in microbial biofilms. Anal Chem 2024, 96, 8308–8316. [Google Scholar] [CrossRef] [PubMed]
- Teschler, J.K.; Nadell, C.D.; Drescher, K.; Yildiz, F.H. Mechanisms underlying Vibrio cholerae biofilm formation and dispersion. Annu Rev Microbiol 2022, 76, 503–532. [Google Scholar] [CrossRef]
- Shrestha, P.; Razvi, A.; Fung, B.L.; Eichinger, S.J.; Visick, K.L. Mutational analysis of Vibrio fischeri c-di-GMP-modulating genes reveals complex regulation of motility. J Bacteriol 2022, 204. [Google Scholar] [CrossRef]
- Ragupathi, H.; Pushparaj, M.M.; Gopi, S.M.; Govindarajan, D.K.; Kandaswamy, K. Biofilm matrix: a multifaceted layer of biomolecules and a defensive barrier against antimicrobials. Arch Microbiol 2024, 206, 432. [Google Scholar] [CrossRef]
- Almatroudi, A. Biofilm resilience: molecular mechanisms driving antibiotic resistance in clinical contexts. Biology (Basel) 2025, 14, 165. [Google Scholar] [CrossRef]
- Roilides, E.; Simitsopoulou, M.; Katragkou, A.; Walsh, T.J. How biofilms evade host defenses. Microbiol Spectr 2015, 3. [Google Scholar] [CrossRef]
- Hitzler, S.U.J.; Fernández-Fernández, C.; Montaño, D.E.; Dietschmann, A.; Gresnigt, M.S. Microbial adaptive pathogenicity strategies to the host inflammatory environment. FEMS Microbiol Rev 2025, 49. [Google Scholar] [CrossRef]
- Tanveer, M.; Ntakiyisumba, E.; Won, G. Revealing antimicrobial resistance profile and associated factors of Vibrio vulnificus isolated from clinical, environmental, and seafood samples across Asia: a systematic review and meta-analysis. Heliyon 2024, 10, e40334. [Google Scholar] [CrossRef]
- Azeem, K.; Fatima, S.; Ali, A.; Ubaid, A.; Husain, F.M.; Abid, M. Biochemistry of bacterial biofilm: insights into antibiotic resistance mechanisms and therapeutic intervention. Life 2025, 15, 49. [Google Scholar] [CrossRef] [PubMed]
- Mandel, M.J.; Dunn, A.K. Impact and influence of the natural Vibrio-squid symbiosis in understanding bacterial–animal interactions. Front Microbiol 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Muthulakshmi, T.; Mothadaka, M.P. Vibrio vulnificus and its antimicrobial resistance. In Handbook on antimicrobial resistance. In Handbook on antimicrobial resistance; Springer Nature Singapore: Singapore, 2023; pp. 1–18. [Google Scholar]
- Yang, Y.; Yan, J.; Olson, R.; Jiang, X. 2025. [CrossRef]
- Pang, R.; Xie, T.; Wu, Q.; Li, Y.; Lei, T.; Zhang, J.; Ding, Y.; Wang, J.; Xue, L.; Chen, M.; et al. Comparative genomic analysis reveals the potential risk of Vibrio parahaemolyticus isolated from ready-to-eat foods in china. Front Microbiol 2019, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Steenackers, H.P.; Parijs, I.; Foster, K.R.; Vanderleyden, J. Experimental evolution in biofilm populations. FEMS Microbiol Rev 2016, 40, 373–397. [Google Scholar] [CrossRef]
- Norsworthy, A.N.; Visick, K.L. Gimme Shelter: How Vibrio fischeri successfully navigates an animal’s multiple environments. Front Microbiol 2013, 4. [Google Scholar] [CrossRef]
- Kuper, T.J.; Islam, M.M.; Peirce-Cottler, S.M.; Papin, J.A.; Ford, R.M. Spatial transcriptome-guided multi-scale framework connects Psuedomonas aeruginosa metabolic states to oxidative stress biofilm microenvironment. PLoS Comput Biol 2024, 20, e1012031. [Google Scholar] [CrossRef]
- Lee, H.H.; Ostrov, N.; Wong, B.G.; Gold, M.A.; Khalil, A.S.; Church, G.M. Functional genomics of the rapidly replicating bacterium Vibrio natriegens by CRISPRi. Nat Microbiol 2019, 4, 1105–1113. [Google Scholar] [CrossRef]
- Pipes, B.L.; Nishiguchi, M.K. Nocturnal acidification: a coordinating cue in the Euprymna scolopes–Vibrio fischeri symbiosis. Int J Mol Sci 2022, 23, 3743. [Google Scholar] [CrossRef]
- Sampaio, A.; Silva, V.; Poeta, P.; Aonofriesei, F. Vibrio spp.: life strategies, ecology, and risks in a changing environment. Diversity (Basel) 2022, 14, 97. [Google Scholar] [CrossRef]
- Vezzulli, L.; Pezzati, E.; Brettar, I.; Höfle, M.; Pruzzo, C. Effects of global warming on Vibrio ecology. Microbiol Spectr 2015, 3. [Google Scholar] [CrossRef]
- Sauer, K. Cyclic di-GMP and the regulation of biofilm dispersion. In Microbial Cyclic Di-Nucleotide Signaling; Springer International Publishing: Cham, 2020; pp. 545–560. [Google Scholar]
- Hussa, E.A.; Darnell, C.L.; Visick, K.L. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J Bacteriol 2008, 190, 4576–4583. [Google Scholar] [CrossRef] [PubMed]
- Griend, J.A. Vander; Isenberg, R.Y.; Kotla, K.R.; Mandel, M.J. Transcriptional pathways across colony biofilm models in the symbiont. Vibrio fischeri, 2023. [Google Scholar]
- Youngblom, M.A.; Smith, T.M.; Murray, H.J.; Pepperell, C.S. Adaptation of the mycobacterium tuberculosis transcriptome to biofilm growth. PLoS Pathog 2024, 20, e1012124. [Google Scholar] [CrossRef] [PubMed]
- León, M.; Kokkari, C.; García, K.; Castillo, D.; Katharios, P.; Bastías, R. Diversification of Vibrio anguillarum driven by the bacteriophage CHOED. Front Microbiol 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Hölscher, T.; Dragoš, A.; Cooper, V.S.; Kovács, Á.T. Laboratory evolution of microbial interactions in bacterial biofilms. J Bacteriol 2016, 198, 2564–2571. [Google Scholar] [CrossRef]
- Barraud, N.; Kjelleberg, S.; Rice, S.A. Dispersal from microbial biofilms. Microbiol Spectr 2015, 3. [Google Scholar] [CrossRef]
- Edel, M.; Horn, H.; Gescher, J. Biofilm systems as tools in biotechnological production. Appl Microbiol Biotechnol 2019, 103, 5095–5103. [Google Scholar] [CrossRef]
- de Oliveira, N.S.; da Silva Ramos, R.C.P.; de Paula, R.C.; da Costa Pereira, M.G.; Rosa, R.T.; Bianchini, L.F.; Rosa, E.A.R. Advantages of using biofilms to obtain high-value molecules by microbial biotransformations. Explor Drug Sci 2025. [Google Scholar] [CrossRef]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol 2017, 15, 740–755. [Google Scholar] [CrossRef]
- Ruby, E.G.; Lee, K.-H. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl Environ Microbiol 1998, 64, 805–812. [Google Scholar] [CrossRef]
- Prentice, J.A.; Bridges, A.A.; Bassler, B.L. Synergy between c-di-GMP and quorum-sensing signaling in Vibrio cholerae biofilm morphogenesis. J Bacteriol 2022, 204. [Google Scholar] [CrossRef]
- Kilic, T. Factors affecting biofilm formation and the effects of these factors on bacteria. In Exploring Bacterial Biofilms; IntechOpen, 2025. [Google Scholar]
- Naga, N.G.; El-Badan, D.E.; Ghanem, K.M.; Shaaban, M.I. It is the time for quorum sensing inhibition as alternative strategy of antimicrobial therapy. Cell Communication and Signaling 2023, 21, 133. [Google Scholar] [CrossRef] [PubMed]
- Yun, M.S.; Sun, J.; Lovejoy, C.; Lee, S.H. Editorial: Microbial response to a rapidly changing marine environment: global warming and ocean acidification. Front Microbiol 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Portas, A.; Carriot, N.; Barry-Martinet, R.; Ortalo-Magné, A.; Hajjoul, H.; Dormoy, B.; Culioli, G.; Quillien, N.; Briand, J.-. François. Shear stress controls prokaryotic and eukaryotic biofilm communities together with eps and metabolomic expression in a semi-controlled coastal environment in the NW Mediterranean sea. Environ Microbiome 2024, 19, 109. [Google Scholar] [CrossRef]
- Chan, W.Y.; Rudd, D.; van Oppen, M.J. Spatial metabolomics for symbiotic marine invertebrates. Life Sci Alliance 2023, 6, e202301900. [Google Scholar] [CrossRef] [PubMed]
- Yeor-Davidi, E.; Zverzhinetsky, M.; Krivitsky, V.; Patolsky, F. Real-time monitoring of bacterial biofilms metabolic activity by a redox-reactive nanosensors array. J Nanobiotechnology 2020, 18, 81. [Google Scholar] [CrossRef]
- Yan, X.; Liao, H.; Wang, C.; Huang, C.; Zhang, W.; Guo, C.; Pu, Y. An improved bacterial single-cell RNA-seq reveals biofilm heterogeneity. Elife 2024, 13. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




