Submitted:
02 June 2023
Posted:
06 June 2023
Read the latest preprint version here
Abstract

Keywords:
1. Introduction
2. Results
2.1. Plastid Genome Features
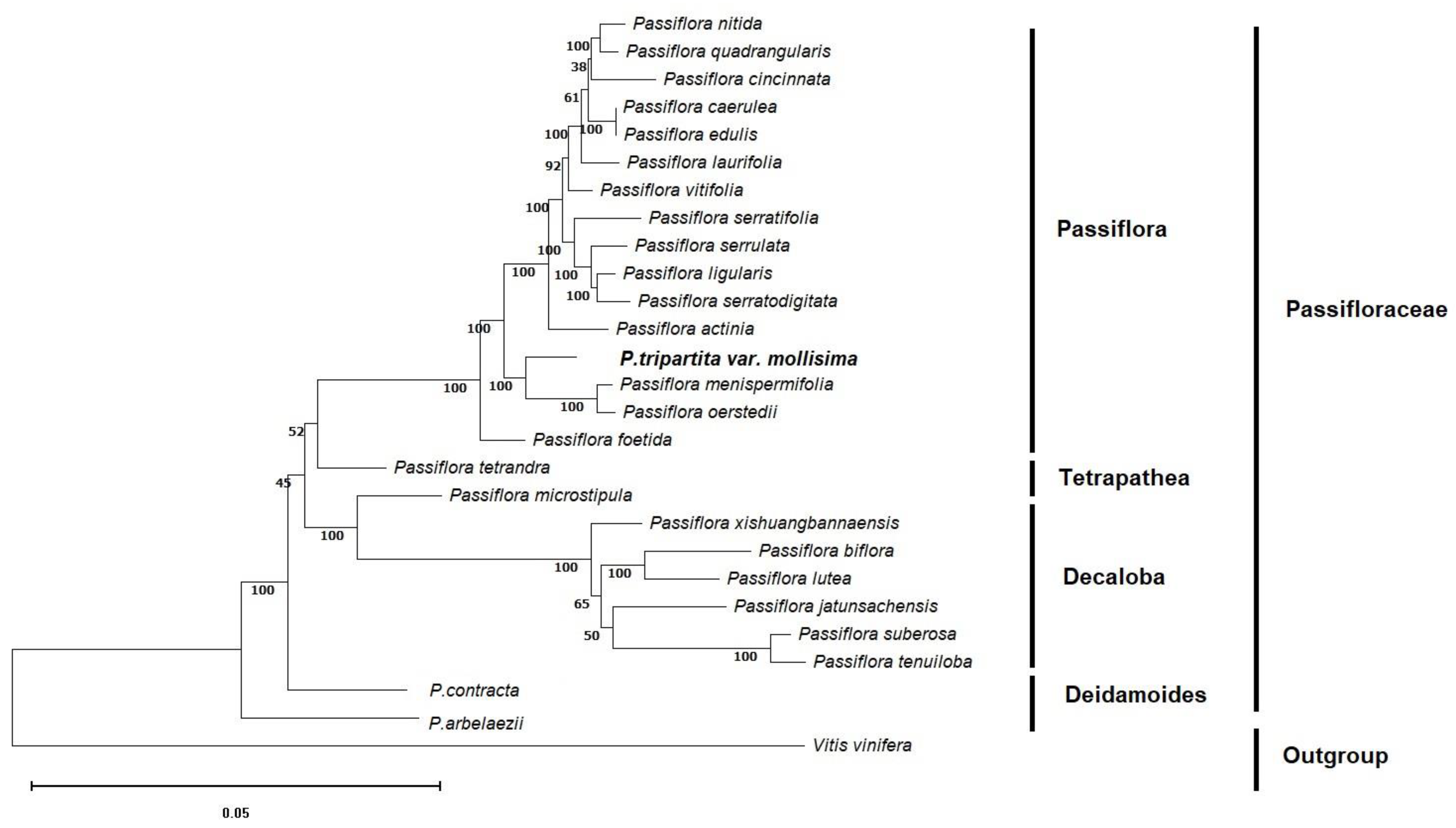
2.2. Phylogenetic Analysis
3. Discussion
3.1. Plastome of Passiflora tripartite var. mollissima
3.2. Phylogenetic Reconstruction of P. tripartita var. mollissima
4. Materials and Methods
4.1. Plant Material
4.2. DNA Extraction
4.3. Genome Sequencing, Assembly, and Annotation
4.3.1. Sequencing
4.3.2. Assembly
4.3.3. Annotation
4.4. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ITIS. (2022) Passiflora tripartita var. mollisima (Kunth) Holms-Niels. & P.M. Jørg. Available online: https://www.itis.gov/ (accessed on 1 Jun 2023).
- Primot, S.; Coppens D’Eeckenbrugge, G.; Rioux, V.; Ocampo, J. A.; Garcin, F. Variación morfológica de tres especies de curubas (Passiflora tripartita var. mollissima, P. tarminiana y P. mixta) y sus híbridos en el Valle del Cauca (Colombia). Rev. Bras. Frutic. 2005, 27, 467–471. [Google Scholar] [CrossRef]
- Mayorga, M.; Fischer, G.; Melgarejo, L. M.; Parra-Coronado, A. Growth, development and quality of Passiflora tripartita var. mollissima fruits under two environmental tropical conditions. J. Appl. Bot. Food Qual. 2020, 93, 66–75. [Google Scholar] [CrossRef]
- Coppens D’Eeckenbrugge, G.; Conservación y utilización de recursos geneticos de pasifloras. COLCIENCIAS Informe final 1999-2001. Available online: https://agritrop.cirad.fr/576977 (accessed on 01 Jun 2023).
- Ocampo, J.; Coppens d’Eeckenbrugge, G.C. Morphological characterization in the genus passiflora l.: an approach to understanding its complex variability. Plant Syst Evol. 2017, 303, 531–558. [Google Scholar] [CrossRef]
- Segura, S.D.; D’Eeckenbrugge, G.C.; Ocampo, C.H.; Ollitrault, P. Isozyme variation in passiflora subgenus tacsonia: Geographic and interspecific differentiation among the three most common species. Genet. Resour. Crop Evol. 2005, 52, 455–463. [Google Scholar] [CrossRef]
- Tapia, M. E; Fries, A. M. Guía de campo de los cultivos andinos, 1st ed.; FAO: Roma, Italy, 2007; pp. 22–114. ISBN 978-92-5-305682-8. [Google Scholar]
- Ríos-García, G. Nivel de aceptabilidad del vino de tumbo serrano (Passiflora mollissima) elaborado con los parámetros tecnológicos óptimos en la ciudad de Huánuco 2015. Master thesis, Universidad Nacional Hermilio Valdizán de Huánuco, Huánuco, 06 November.
- Chaparro-Rojas, D.C.; Maldonado, M.E.; Franco-Londoño, M.C.; Urango-Marchena, L.A. Características nutricionales y antioxidantes de la fruta curuba larga (Passifora mollissima Bailey). Perspect. Nutr. Hum. 2014, 16, 203–212. [Google Scholar] [CrossRef]
- Leterme, P.; Buldgen, A.; Estrada, F.; Londoño, A.M. Mineral content of tropical fruits and unconventional foods of the Andes and the rain forest of Colombia. Food. Chem. 2006, 95, 644–652. [Google Scholar] [CrossRef]
- Giambanelli, E.; Gómez-Caravaca, A.M.; Ruiz-Torralba, A.; Guerra-Hernández, E.J.; Figueroa-Hurtado, J.G.; García-Villanova, B.; Verardo, V. New advances in the determination of free and bound phenolic compounds of banana passion fruit pulp (Passiflora tripartita, var. mollissima (kunth) l.h. bailey) and their in vitro antioxidant and hypoglycemic capacities. Antioxidants, 2020, 9, 628. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Jian, Y.; Liu, Y.; Jiang, S.; Muhammad, D.; Wang, W. Flavanols from nature: A phytochemistry and biological activity review. Molecules, 2022, 27, 719. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol., 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Dobrogojski, J.; Adamiec, M.; Lucinski, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Ozeki, H.; Umesono, K.; Inokuchi, H.; Kohchi, T.; Ohyama, K. The chloroplast genome of plants: a unique origin. Genome. 1989, 31, 169–174. [Google Scholar] [CrossRef]
- Wang, W.; Lanfear, R. Long-reads reveal that the chloroplast genome exists in two distinct versions in most plants. Genome Biol Evol. 2019, 11, 3372–3381. [Google Scholar] [CrossRef]
- Marks, R.A.; Hotaling, S.; Frandsen, P.B.; VanBuren, R. Representation and participation across 20 years of plant genome sequencing. Nat Plants. 2021, 7, 1571–1578. [Google Scholar] [CrossRef]
- Sun, Y.; Shang, L.; Zhu, Q.H.; Fan, L.; Guo, L. Twenty years of plant genome sequencing: achievements and challenges. Trends Plant Sci. 2022, 27, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Cauz-Santos, L.A.; Munhoz, C.F.; Rodde, N.; Cauet, S.; Santos, A.A.; Penha, H.A.; Dornelas, M.C.; Varani, A.M.; Oliveira, G.C.X.; Gergès, H.; Vieira, M.L.C. The Chloroplast Genome of Passiflora edulis (Passifloraceae) Assembled from Long Sequence Reads: Structural Organization and Phylogenomic Studies in Malpighiales. Front Plant Sci. 2017, 8, 334. [Google Scholar] [CrossRef] [PubMed]
- Hao, C.; Wu, F. The complete chloroplast genome sequence of Passiflora xishuangbannaensis (Passifloraceae), a vine endemic to Yunnan, China. Mitochondrial DNA B Resour. 2021, 6, 1763–1764. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.F.; Ni, S.B.; Liu, S.H.; Liu, J. The complete chloroplast genome of Passiflora caerulea, a tropical fruit with a distinctive aroma. Mitochondrial DNA B Resour. 2021, 6, 488–490. [Google Scholar] [CrossRef] [PubMed]
- Mou, H.F.; Huang, W.H.; Liu, J.Y.; Wen, F.; Tian, Q.L.; Wu, Y.Y.; Fu, L.F.; Zhang, Y.J.; Wei, Y.G. Complete chloroplast genome sequence of Passiflora serrulata Jacq. (Passifloraceae). Mitochondrial DNA B Resour. 2021, 6, 191–193. [Google Scholar] [CrossRef] [PubMed]
- Hopley, T.; Webber, B.L.; Raghu, S.; Morin, L.; Byrne, M. Revealing the Introduction History and Phylogenetic Relationships of Passiflora foetida sensu lato in Australia. Front Plant Sci. 2021, 12, 651805. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, B.; Weng, M.L.; Theriot, E.C.; Gilbert, L.E.; Ruhlman, T.A.; Krosnick, S.E.; Jansen, R.K. Highly accelerated rates of genomic rearrangements and nucleotide substitutions in plastid genomes of Passiflora subgenus Decaloba. Mol. Phylogenet. Evol. 2019, 138, 53–64. [Google Scholar] [CrossRef]
- Gioppato, H.A.; da Silva, M.B.; Carrara, S.; Palermo, B.R.Z.; de Souza Moraes, T.; Dornelas, M.C. Genomic and transcriptomic approaches to understand Passiflora physiology and to contribute to passionfruit breeding. Theor. Exp. Plant Physiol. 2019, 31, 173–181. [Google Scholar] [CrossRef]
- Ohyama, K.; Fukuzawa, H.; Kohchi, T.; Shirai, H.; Sano, T.; Sano, S.; Umesono, K.; Shiki, Y. , Takeuchi, M., Chang, Z., Aota, S.I., Inokuchi, H.; Ozeki, H. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986, 322, 572–574. [Google Scholar] [CrossRef]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayashida, N.; Matsubayashi, T.; Zaita, N.; Chunwongse, J.; Obokata, J.; Yamaguchi-Shinozaki, K.; Ohto, C.; Torazawa, K.; Meng, B. Y.; Sugita, M.; Deno, H.; Kamogashira, T.; Yamada, K.; Kusuda, J.; Takaiwa, F.; Kato, A.; Tohdoh, N.; Shimada, H.; Sugiura, M. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J., 1986, 5, 2043–2049. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Nguyen, T.N.L.; Doan, T.N.; Nguyen, T.T.N.; Phạm, M.H.; Le, T.L.; Sy, D.T.; Chu, H.H.; Chu, H.M. Complete chloroplast genome of novel Adrinandra megaphylla Hu species: molecular structure, comparative and phylogenetic analysis. Sci. Rep. 2021, 11, 11731. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Poczai, P.; Hyvönen, J.; Tang, J.; Amiryousefi, A. Chloroplot: An Online Program for the Versatile Plotting of Organelle Genomes. Front. Genet. 2020, 11, 576124. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, S.; Bédard, J.; Hirano, M.; Hirabayashi, Y.; Oishi, M.; Imai, M.; Takase, M.; Ide, T.; Nakai, M. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science. 2013, 339, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, S.; Asakura, Y.; Imai, M.; Nakahira, Y.; Kotani, Y.; Hashiguchi, Y.; Nakai, Y.; Takafuji, K.; Bédard, J.; Hirabayashi-Ishioka, Y.; Mori, H. , Shiina, T.; Nakai, M. A Ycf2-FtsHi Heteromeric AAA-ATPase Complex Is Required for Chloroplast Protein Import. Plant Cell. 2018, 30, 2677–2703. [Google Scholar] [CrossRef]
- Gabaldón, T. Evolution of proteins and proteomes: a phylogenetics approach. Evol. Bioinform. Online. 2005, 1, 51–61. [Google Scholar] [CrossRef]
- Zhang, N.; Zeng, L.; Shan, H.; Ma, H. Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms. New Phytol. 2012, 195, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, T.G.; Lopes, A.S.; Welter, J.F.; Yotoko, K.S.C.; Otoni, W.C.; Vieira, L.D.N.; Guerra, M.P.; Nodari, R.O.; Balsanelli, E.; Pedrosa, F.O.; de Souza, E.M.; Rogalski, M. Plastome sequences of the subgenus Passiflora reveal highly divergent genes and specific evolutionary features. Plant Mol. Biol. 2020, 104, 21–37. [Google Scholar] [CrossRef]
- Cauz-Santos, L.A.; da Costa, Z.P.; Callot, C.; Cauet, S.; Zucchi, M.I.; Bergès, H.; van den Berg, C.; Vieira, M.L.C. A Repertory of Rearrangements and the Loss of an Inverted Repeat Region in Passiflora Chloroplast Genomes. Genome Biol. Evol. 2020, 12, 1841–1857. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Modi, A.; Vai, S.; Caramelli, D. , M. The illumina sequencing protocol and the novaseq 6000 system. In Bacterial Pangenomics: Methods and Protocols, 2nd edn; Mengoni, A., Bacci, G., Fondi, M., Eds.; Springer: New York, USA, 2021; Volume 2242, pp. 15–42. ISBN 978-1-0716-1099-2. [Google Scholar]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Res. 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, H.; Jiang, M. , Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]

| Features | Poro-poro 1 |
|---|---|
| Genome size (bp) | 163,451 |
| LSC length (bp) | 85, 525 |
| SSC length (bp) | 13,518 |
| IR length (bp) | 32,204 |
| Total GC content (%) | 36.87 |
| A content (%) | 30.79 |
| T(U) content (%) | 32.34 |
| G content (%) | 18.20 |
| C content (%) | 18.67 |
| Total number of genes | 128 |
| Protein-coding genes | 84 |
| rRNA coding genes | 7 |
| tRNA coding genes | 37 |
| Genes duplicated in IR regions | 17 |
| Total introns | 14 |
| Single introns (gene) | 12 |
| Double introns (gene) | 2 |
| Group of genes | Gene names |
|---|---|
| Photosystem I | psaA, psaB, psaC, psaI, psaJ, ycf3 **, ycf4 |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| Cytochrome b/f complex | petA, petB, petD *, petG, petL, petN |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI |
| NADH dehydrogenase | ndhA*, ndhB * (X2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| RubisCO large subunit | rbcL |
| DNA-dependent RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 |
| Ribosomal proteins (SSU) | rps2, rps3, rps4, rps8, rps11, rps12 ** (X2), rps14, rps15, rps16, rps18, rps19 (X2) |
| Ribosomal proteins (LSU) | rpl2 * (X2), rpl14, rpl16 *, rpl20, rpl22, rpl23 (X2), rpl32, rpl33, rpl36 |
| Acetyl-CoA carboxylase | accD |
| C-type cytochrome synthesis | ccsA |
| Envelope membrane protein | cemA |
| Protease | clpP |
| Translational initiation factor IF-1 | infA |
| Maturase | matK |
| Component of TIC complex | yct1, ycf2 |
| Unknown function protein-coding | ycf15 (X2) |
| Ribosomal RNAs | rrn4.5, rrn5 (X2), rrn16 (X2), rrn23 (X2) |
| Transfer RNAs | trnA-UGC * (X2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC *, trnH-GUG, trnI-CAU (X2), trnI-GAU * (X2), trnK-UUU *, trnL-CAA (X2), trnL-UAA *, trnL-UAG, trnM-CAU (X2), trnN-GUU (X2), trnP-UGG, trnQ-UUG, trnR-ACG (X2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC (X2), trnV-UAC *, trnW-CCA, trnY-GUA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





