Submitted:
23 October 2024
Posted:
24 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Animals, Diets, Blood and Urine Samples
2.2. Metabolite Analysis Protocol
2.3. Organic Acid Analysis Procedure
2.4. FIA/LC—MS/MS Analysis Method
2.5. Statistical Analysis
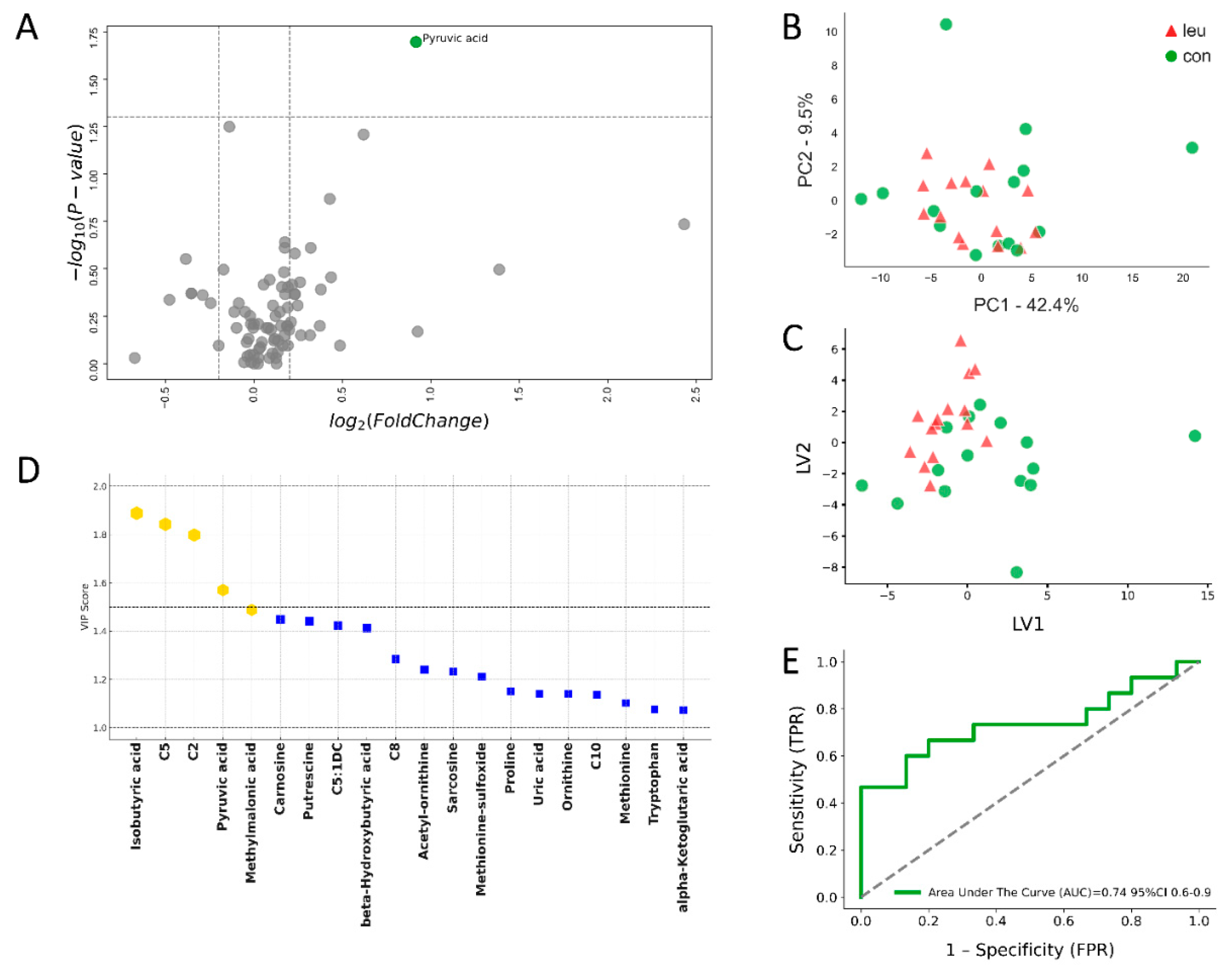
3. Results
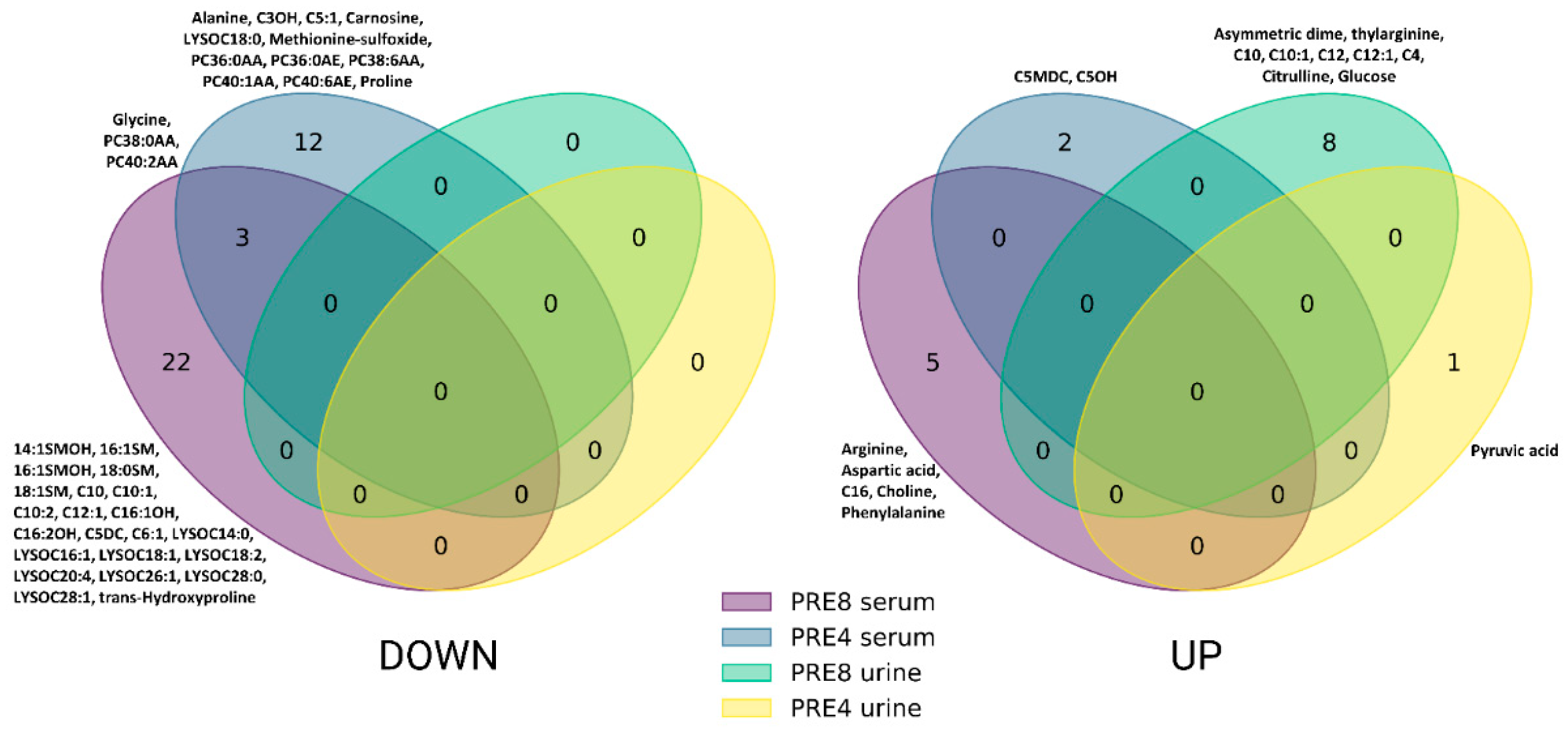
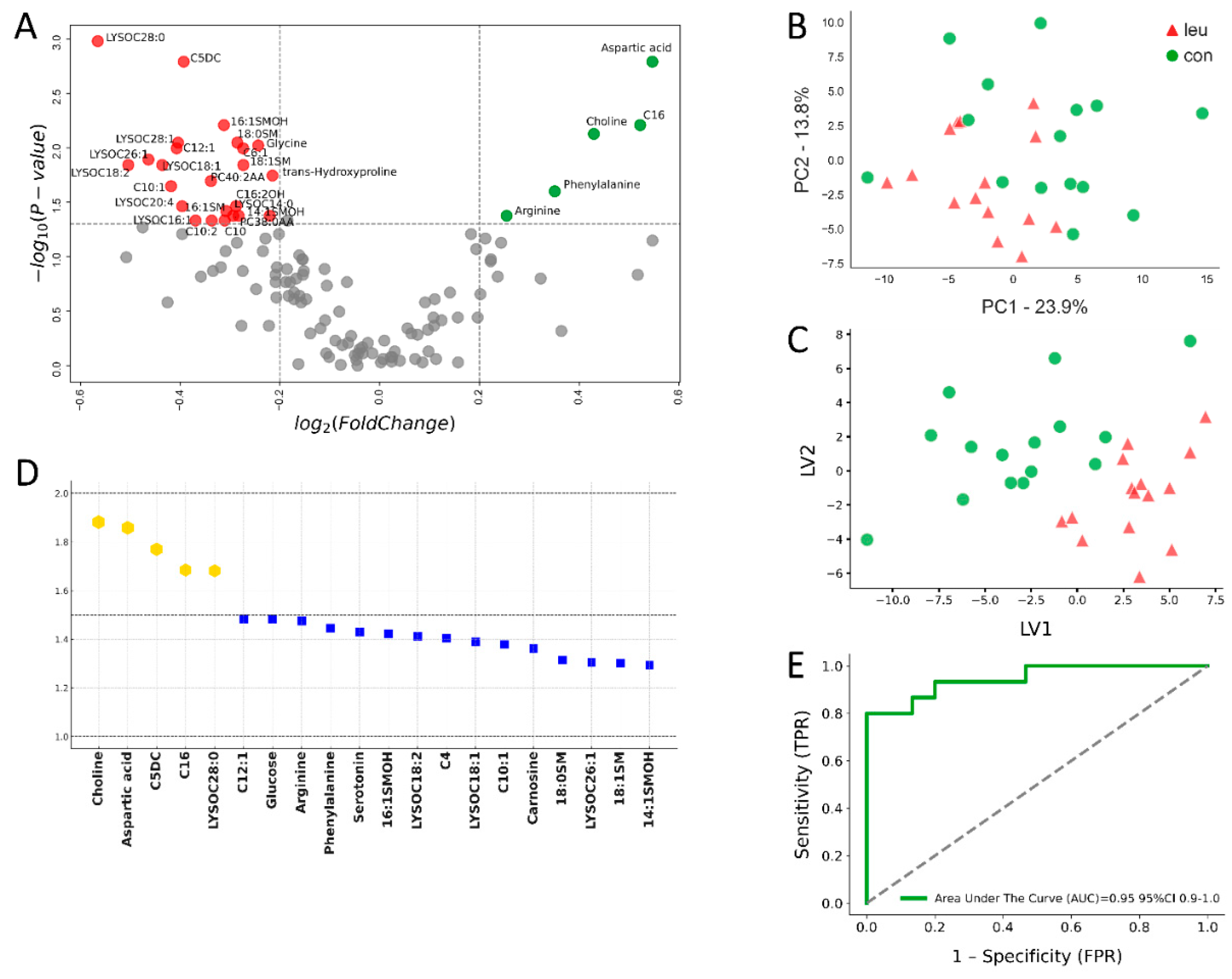
3.1. Blood Metabolomics Alterations in Leukemic Cows During Dry off Period
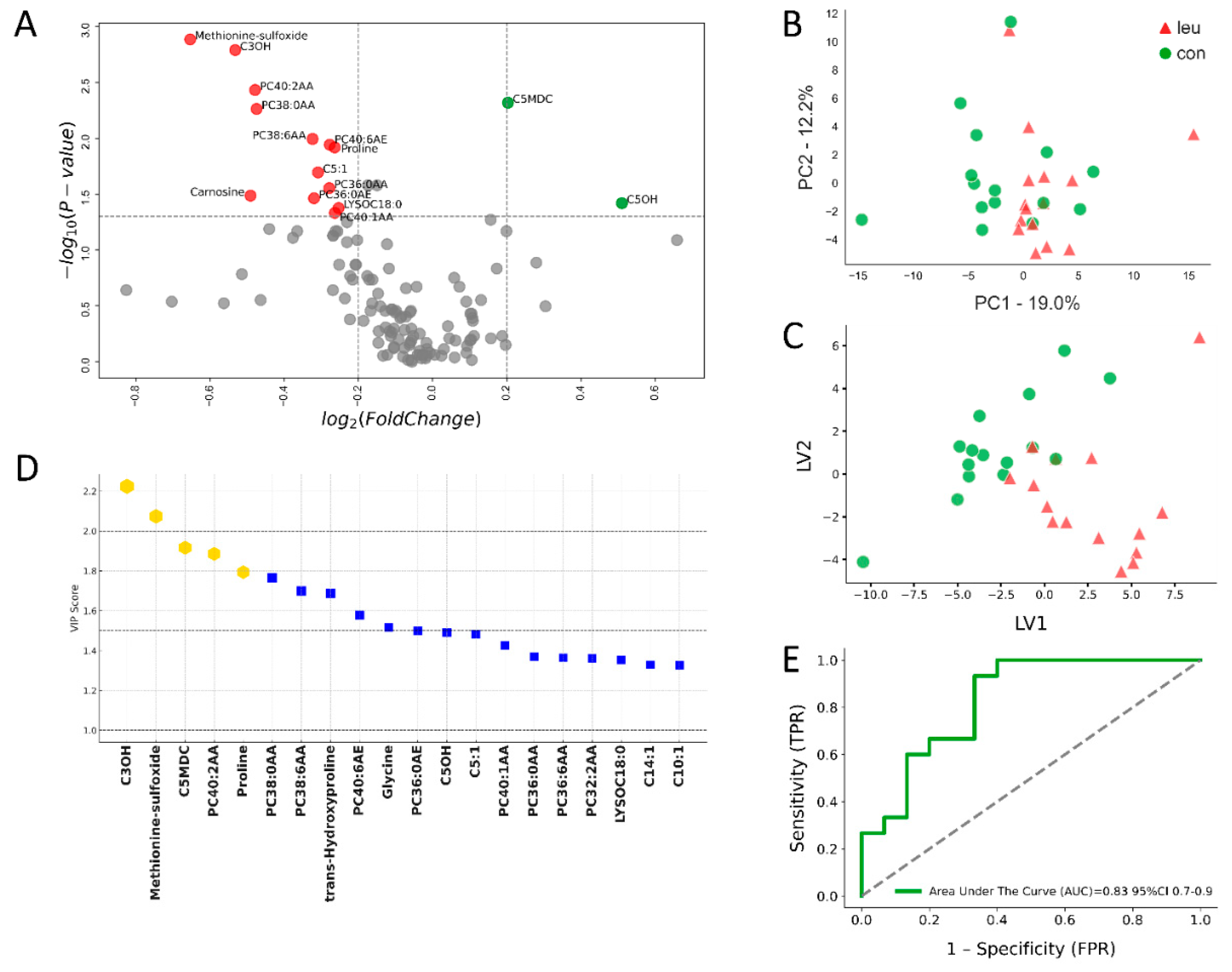
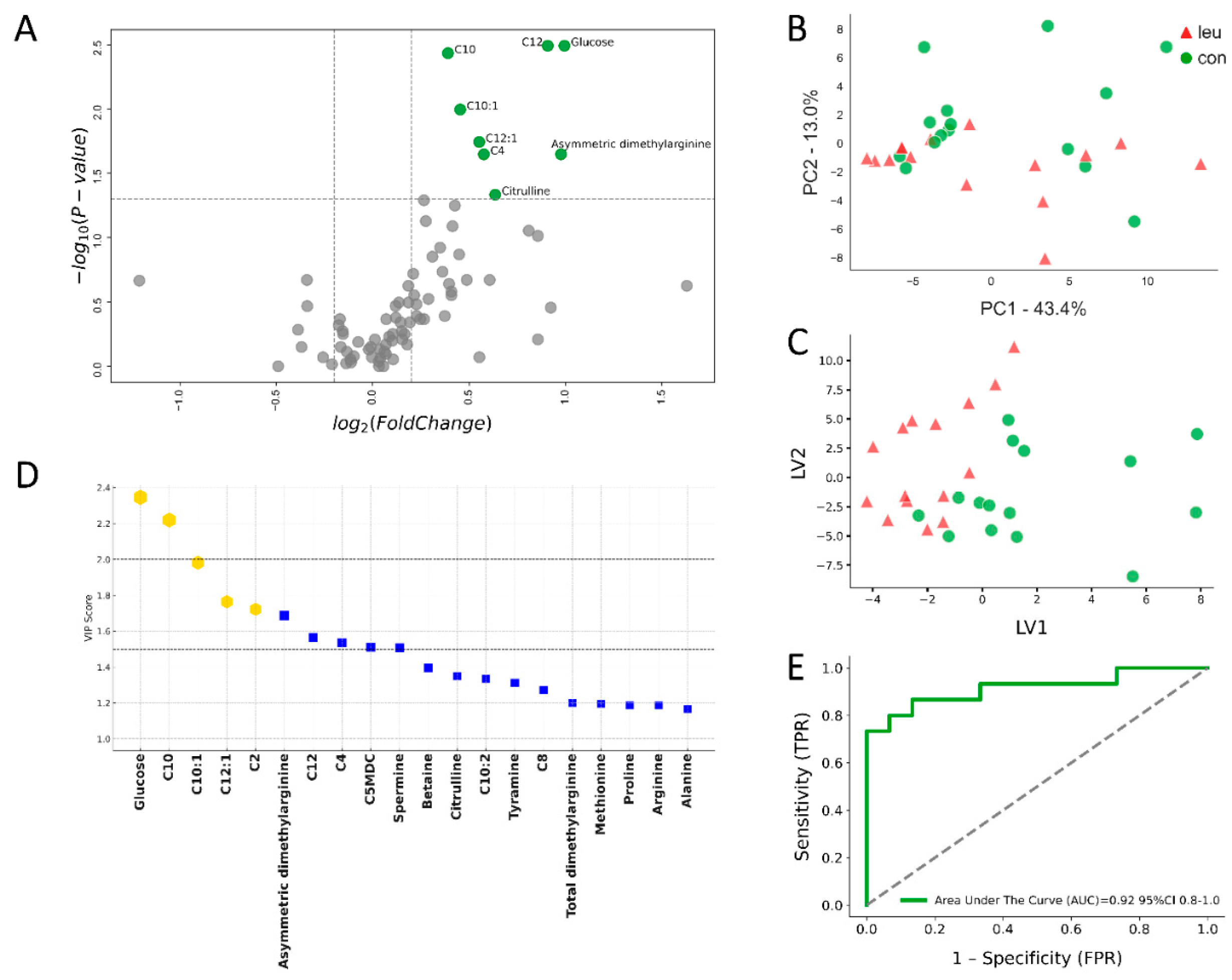
3.2. Urinary Metabolomics Alterations in Leukosis Cows During the Dry off Period
4. Discussion
4.1. Blood Alterations in BLV-Infected Cows During the Dry off Period
4.2. Urinary Metabolite Alterations in BLV-Infected Cows During the Dry off Period
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuczewski, A.; Hogeveen, H.; Orsel, K.; Wolf, R.; Thompson, J.; Spackman, E.; van der Meer, F. Economic evaluation of 4 bovine leukemia virus control strategies for Alberta dairy farms. J. Dairy Sci. 2019, 102, 2578–2592. [Google Scholar] [CrossRef] [PubMed]
- LaDronka, R.M.; Ainsworth, S.; Wilkins, M.J.; Norby, B.; Byrem, T.M.; Bartlett, P.C. Prevalence of bovine leukemia virus antibodies in US dairy cattle. Vet. Med. Int. 2018, 2018, 5831278. [Google Scholar] [CrossRef] [PubMed]
- Gillet, N.; Florins, A.; Boxus, M.; Burteau, C.; Nigro, A.; Vandermeers, F.; Balon, H.; Bouzar, A.-B.; Defoiche, J.; Burny, A. Mechanisms of leukemogenesis induced by bovine leukemia virus: Prospects for novel anti-retroviral therapies in human. Retrovirology 2007, 4, 32. [Google Scholar] [CrossRef] [PubMed]
- Goff, S.P. Retroviridae: The retroviruses and their replication. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2001. [Google Scholar]
- Schambow, R.; Poulsen, K.; Bolin, S.; Krahn, D.; Norby, B.; Sockett, D.; Ruegg, P. Apparent prevalence of Mycoplasma wenyonii, Candidatus Mycoplasma haemobos, and bovine leukemia virus in Wisconsin and Michigan dairy cattle herds. JDS Commun. 2021, 2, 61–66. [Google Scholar] [CrossRef]
- Bartlett, P.C.; Ruggiero, V.J.; Hutchinson, H.C.; Droscha, C.J.; Norby, B.; Sporer, K.R.; Taxis, T.M. Current developments in the epidemiology and control of enzootic bovine leukosis as caused by bovine leukemia virus. Pathogens 2020, 9, 1058. [Google Scholar] [CrossRef]
- Tomiyasu, T.; Mori, H.; Okazaki, K. Epidemiological evidence for early-onset of enzootic bovine leukosis by L233-Tax-carrying bovine leukemia virus in Japanese Black cattle. J. Vet. Med. Sci. 2022, 84, 1216–1220. [Google Scholar] [CrossRef]
- Kuczewski, A.; Orsel, K.; Barkema, H.W.; Mason, S.; Erskine, R.; van der Meer, F. Invited review: Bovine leukemia virus—Transmission, control, and eradication. J. Dairy Sci. 2021, 104, 6358–6375. [Google Scholar] [CrossRef]
- Rodríguez, S.M.; Florins, A.; Gillet, N.; de Brogniez, A.; Sánchez-Alcaraz, M.T.; Boxus, M.; Boulanger, F.; Gutiérrez, G.; Trono, K.; Alvarez, I.; et al. Preventive and therapeutic strategies for bovine leukemia virus: Lessons for HTLV. Viruses 2011, 3, 1210–1248. [Google Scholar] [CrossRef]
- Buehring, G.C.; DeLaney, A.; Shen, H.; Chu, D.L.; Razavian, N.; Schwartz, D.A.; Demkovich, Z.R.; Bates, M.N. Bovine leukemia virus discovered in human blood. BMC Infect. Dis. 2019, 19, 1–10. [Google Scholar] [CrossRef]
- Szczotka, M.; Kuźmak, J. Expression of bovine leukaemia virus (BLV) gp51 protein in blood and milk cells of cows with leukosis. J. Vet. Res. 2022, 66, 305–315. [Google Scholar] [CrossRef]
- Notsu, K.; Wiratsudakul, A.; Mitoma, S.; El Daous, H.; Kaneko, C.; El-Khaiat, H.M.; Norimine, J.; Sekiguchi, S. Quantitative risk assessment for the introduction of bovine leukemia virus-infected cattle using a cattle movement network analysis. Pathogens 2020, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Evermann, J.F.; DeAvila, D.M.; Parish, S.M.; Merritt, C.H.; Noble, K.C.; Srikanth, S.; Bronowski, A.L.E. Evaluation of a serum ELISA for detection of bovine leukemia viral antibodies in milk samples. J. Vet. Diagn. Investig. 2019, 31, 598–600. [Google Scholar] [CrossRef] [PubMed]
- Borjigin, L.; Yoneyama, S.; Saito, S.; Polat, M.; Inokuma, M.; Shinozaki, Y.; Tanaka, N.; Yamanaka, R.; Yasui, A.; Mimura, M.; et al. A novel real time PCR assay for bovine leukemia virus detection using mixed probes and degenerate primers targeting novel BLV strains. J. Virol. Methods 2021, 297, 114264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Tobolski, D.; Zwierzchowski, G.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. Identification of serum-predictive biomarkers for subclinical mastitis in dairy cows and new insights into the pathobiology of the disease. J. Agric. Food Chem. 2022, 70, 1724–1746. [Google Scholar] [CrossRef]
- Zhang, G.; Tobolski, D.; Zwierzchowski, G.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. A targeted serum metabolomics GC-MS approach identifies predictive blood biomarkers for retained placenta in Holstein dairy cows. Metabolites 2021, 11, 633. [Google Scholar] [CrossRef]
- Watanabe, A.; Murakami, H.; Kakinuma, S.; Murao, K.; Oomae, K.; Akamatsu, H.; Seto, T.; Shinozuka, Y.; Kawai, K. Predicting an increased risk of severe clinical mastitis and economic loss using a threshold value of bovine leukemia virus proviral load. Am. J. Vet. Res. 2023, 1, AOP. [Google Scholar] [CrossRef]
- Sandev, N.; Koleva, M.; Binev, R.; Ilieva, D. Influence of enzootic bovine leukosis virus upon the incidence of subclinical mastitis in cows at a different stage of infection. Vet. Arh. 2004, 74, 411–416. [Google Scholar]
- Bolkun, L.; Pienkowski, T.; Sieminska, J.; Godzien, J.; Pietrowska, K.; Kłoczko, J.; Wierzbowska, A.; Moniuszko, M.; Ratajczak, M.; Kretowski, A. Metabolomic profile of acute myeloid leukaemia parallels prognosis and response to therapy. Sci. Rep. 2023, 13, 21809. [Google Scholar] [CrossRef]
- Finney, R.E.; Nudelman, E.; White, T.; Bursten, S.; Klein, P.; Leer, L.L.; Wang, N.; Waggoner, D.; Singer, J.W.; Lewis, R.A. Pharmacological inhibition of phosphatidylcholine biosynthesis is associated with induction of phosphatidylinositol accumulation and cytolysis of neoplastic cell lines. Cancer Res. 2000, 60, 5204–5213. [Google Scholar]
- Linkous, A.; Yazlovitskaya, E. Cytosolic phospholipase A2 as a mediator of disease pathogenesis. Cell. Microbiol. 2010, 12, 1369–1377. [Google Scholar] [CrossRef]
- Florins, A.-F.; Gillet, N.; Asquith, B.; Boxus, M.; Burteaux, C.; Twizere, J.-C.; Urbain, P.; Vandermeers, F.; Debacq, C.; Sanchez-Alcaraz, M.T. Cell dynamics and immune response to BLV infection: A unifying model. Front. Biosci. 2007, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Sharma-Walia, N. Curbing lipids: Impacts on cancer and viral infection. Int. J. Mol. Sci. 2019, 20, 644. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Pokrovsky, V.S.; Ivanova-Radkevich, V.I.; Kuznetsova, O.M. Sphingolipid metabolism in tumor cells. Biochem. (Moscow) 2023, 88, 847–866. [Google Scholar] [CrossRef] [PubMed]
- Nekoei, S.; Hafshejani, T.T.; Doosti, A.; Khamesipour, F. Molecular detection of bovine leukemia virus in peripheral blood of Iranian cattle, camel and sheep. Pol. J. Vet. Sci. 2015, 18, 301–305. [Google Scholar] [CrossRef]
- Bensinger, S.J.; Bradley, M.N.; Joseph, S.B.; Zelcer, N.; Janssen, E.M.; Hausner, M.A.; Shih, R.; Parks, J.S.; Edwards, P.A.; Jamieson, B.D. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell 2008, 134, 97–111. [Google Scholar] [CrossRef]
- Jantscheff, P.; Schlesinger, M.; Fritzsche, J.; Taylor, L.A.; Graeser, R.; Kirfel, G.; Fürst, D.O.; Massing, U.; Bendas, G. Lysophosphatidylcholine pretreatment reduces VLA-4 and P-Selectin–mediated B16.F10 melanoma cell adhesion in vitro and inhibits metastasis-like lung invasion in vivo. Mol. Cancer Ther. 2011, 10, 186–197. [Google Scholar] [CrossRef]
- Mehedint, M.G.; Zeisel, S.H. Choline's role in maintaining liver function: New evidence for epigenetic mechanisms. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 339–345. [Google Scholar] [CrossRef]
- Blusztajn, J.K.; Mellott, T.J. Choline nutrition programs brain development via DNA and histone methylation. Cent. Nerv. Syst. Agents Med. Chem. 2012, 12, 82–94. [Google Scholar] [CrossRef]
- Delage, B.; Fennell, D.A.; Nicholson, L.; McNeish, I.; Lemoine, N.R.; Crook, T.; Szlosarek, P.W. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int. J. Cancer 2010, 126, 2762–2772. [Google Scholar] [CrossRef]
- Al-Koussa, H.; El Mais, N.; Maalouf, H.; Abi-Habib, R.; El-Sibai, M. Arginine deprivation: A potential therapeutic for cancer cell metastasis? A review. Cancer Cell Int. 2020, 20, 150. [Google Scholar] [CrossRef] [PubMed]
- Fultang, L.; Vardon, A.; De Santo, C.; Mussai, F. Molecular basis and current strategies of therapeutic arginine depletion for cancer. Int. J. Cancer 2016, 139, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.; Bhaumik, J.; Babykutty, S.; Banerjee, U.; Fukumura, D. Arginine dependence of tumor cells: Targeting a chink in cancer’s armor. Oncogene 2016, 35, 4957–4972. [Google Scholar] [CrossRef] [PubMed]
- Chandel, N.S. Amino acid metabolism. Cold Spring Harb. Perspect. Biol. 2021, 13, a040584. [Google Scholar] [CrossRef]
- Van Gastel, N.; Spinelli, J.B.; Sharda, A.; Schajnovitz, A.; Baryawno, N.; Rhee, C.; Oki, T.; Grace, E.; Soled, H.J.; Milosevic, J. Induction of a timed metabolic collapse to overcome cancer chemoresistance. Cell Metab. 2020, 32, 391–403.e396. [Google Scholar] [CrossRef]
- Holeček, M. Aspartic acid in health and disease. Nutrients 2023, 15, 4023. [Google Scholar] [CrossRef]
- Chandel, N.S. Nucleotide metabolism. Cold Spring Harb. Perspect. Biol. 2021, 13, a040592. [Google Scholar] [CrossRef]
- Pine, M.J. Improved host defense against L1210 leukemia by deprivation of dietary phenylalanine. J. Natl. Cancer Inst. 1981, 66, 783–785. [Google Scholar] [CrossRef]
- Pine, M.J. Effect of low phenylalanine diet on murine leukemia L1210. J. Natl. Cancer Inst. 1978, 60, 633–641. [Google Scholar] [CrossRef]
- McCarty, M.F.; O'Keefe, J.H.; DiNicolantonio, J.J. Dietary glycine is rate-limiting for glutathione synthesis and may have broad potential for health protection. Ochsner J. 2018, 18, 81–87. [Google Scholar]
- Xia, J.; Zhang, J.; Wu, X.; Du, W.; Zhu, Y.; Liu, X.; Liu, Z.; Meng, B.; Guo, J.; Yang, Q. Blocking glycine utilization inhibits multiple myeloma progression by disrupting glutathione balance. Nat. Commun. 2022, 13, 4007. [Google Scholar] [CrossRef] [PubMed]
- Verstraete, P.; De Keersmaecker, K.; Kampen, K.R. Drivers of de novo serine/glycine synthesis in acute leukemia. FEBS Lett. 2023, 597, 2145–2146. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Castillo, A.; Vooijs, M.; Kampen, K.R. Linking serine/glycine metabolism to radiotherapy resistance. Cancers 2021, 13, 1191. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wu, G. Roles of dietary glycine, proline, and hydroxyproline in collagen synthesis and animal growth. Amino Acids 2018, 50, 29–38. [Google Scholar] [CrossRef]
- Graf, M.; Wilson, D.N. Intracellular antimicrobial peptides targeting the protein synthesis machinery. Antimicrob. Pept.: Basic Clin. Appl. 2019, 73–89. [Google Scholar]
- Mishra, A.K.; Choi, J.; Moon, E.; Baek, K.-H. Tryptophan-rich and proline-rich antimicrobial peptides. Molecules 2018, 23, 815. [Google Scholar] [CrossRef]
- Martínez, Y.; Li, X.; Liu, G.; Bin, P.; Yan, W.; Más, D.; Valdivié, M.; Hu, C.-A.A.; Ren, W.; Yin, Y. The role of methionine on metabolism, oxidative stress, and diseases. Amino Acids 2017, 49, 2091–2098. [Google Scholar] [CrossRef]
- Levine, R.L.; Moskovitz, J.; Stadtman, E.R. Oxidation of methionine in proteins: Roles in antioxidant defense and cellular regulation. IUBMB Life 2000, 50, 301–307. [Google Scholar] [CrossRef]
- Ward, N.P.; DeNicola, G.M. Sulfur metabolism and its contribution to malignancy. Int. Rev. Cell Mol. Biol. 2019, 347, 39–103. [Google Scholar]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Wu, G. Amino acids: Metabolism, functions, and nutrition. Amino Acids 2009, 37, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Trachootham, D.; Alexandre, J.; Huang, P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat. Rev. Drug Discov. 2009, 8, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Baye, E.; Ukropcova, B.; Ukropec, J.; Hipkiss, A.; Aldini, G.; de Courten, B. Physiological and therapeutic effects of carnosine on cardiometabolic risk and disease. Amino Acids 2016, 48, 1131–1149. [Google Scholar] [CrossRef]
- Nagai, K.; Suda, T. Immunoregulative effects of carnosine and beta-alanine. Nihon Seirigaku Zasshi 1986, 48, 564–571. [Google Scholar]
- Prakash, M.D.; Fraser, S.; Boer, J.C.; Plebanski, M.; de Courten, B.; Apostolopoulos, V. Anti-cancer effects of carnosine—A dipeptide molecule. Molecules 2021, 26, 1644. [Google Scholar] [CrossRef]
- Li, S.; Gao, D.; Jiang, Y. Function, detection and alteration of acylcarnitine metabolism in hepatocellular carcinoma. Metabolites 2019, 9, 36. [Google Scholar] [CrossRef]
- Anand, S.K.; Tikoo, S.K. Viruses as modulators of mitochondrial functions. Adv. Virol. 2013, 2013, 738794. [Google Scholar] [CrossRef]
- Houten, S.M.; Violante, S.; Ventura, F.V.; Wanders, R.J. The biochemistry and physiology of mitochondrial fatty acid β-oxidation and its genetic disorders. Annu. Rev. Physiol. 2016, 78, 23–44. [Google Scholar] [CrossRef]
- Dambrova, M.; Makrecka-Kuka, M.; Kuka, J.; Vilskersts, R.; Nordberg, D.; Attwood, M.M.; Smesny, S.; Sen, Z.D.; Guo, A.C.; Oler, E. Acylcarnitines: Nomenclature, biomarkers, therapeutic potential, drug targets, and clinical trials. Pharmacol. Rev. 2022, 74, 506–551. [Google Scholar] [CrossRef]
- Ametaj, B.; Zhang, G.; Dervishi, E.; Wishart, D. Urinary metabotyping around parturition indicates consistent metabolite signatures that can be used for monitoring and diagnosing of subclinical mastitis in dairy cows. J. Anim. Sci. 2018, 96 (Suppl 3), 19. [Google Scholar] [CrossRef]
- Zwierzchowski, G.; Zhang, G.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. Mass-spec-based urinary metabotyping around parturition identifies screening biomarkers for subclinical mastitis in dairy cows. Res. Vet. Sci. 2020, 129, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Lyu, C.; Li, Z.; Wang, Q.; Ding, Y.; Wang, Y.; Qiu, Y.; Cui, S.; Guo, D.; Xu, R. High expression of ACOT2 predicts worse overall survival and abnormal lipid metabolism: A potential target for acute myeloid leukemia. J. Healthc. Eng. 2022, 2022, 2669114. [Google Scholar] [CrossRef] [PubMed]
- Enooku, K.; Nakagawa, H.; Fujiwara, N.; Kondo, M.; Minami, T.; Hoshida, Y.; Shibahara, J.; Tateishi, R.; Koike, K. Altered serum acylcarnitine profile is associated with the status of nonalcoholic fatty liver disease (NAFLD) and NAFLD-related hepatocellular carcinoma. Sci. Rep. 2019, 9, 10663. [Google Scholar] [CrossRef]
- Schooneman, M.G.; Vaz, F.M.; Houten, S.M.; Soeters, M.R. Acylcarnitines: Reflecting or inflicting insulin resistance? Diabetes 2013, 62, 1–8. [Google Scholar] [CrossRef]
- Zhelev, Z.; Aoki, I.; Lazarova, D.; Vlaykova, T.; Higashi, T.; Bakalova, R. A “weird” mitochondrial fatty acid oxidation as a metabolic “secret” of cancer. Oxid. Med. Cell. Longev. 2022, 2022, 2339584. [Google Scholar] [CrossRef]
- Fodor, T.; Szántó, M.; Abdul-Rahman, O.; Nagy, L.; Dér, Á.; Kiss, B.; Bai, P. Combined treatment of MCF-7 cells with AICAR and methotrexate arrests cell cycle and reverses Warburg metabolism through AMP-activated protein kinase (AMPK) and FOXO1. PLoS ONE 2016, 11, e0150232. [Google Scholar] [CrossRef]
- Ganti, S.; Taylor, S.L.; Kim, K.; Hoppel, C.L.; Guo, L.; Yang, J.; Evans, C.; Weiss, R.H. Urinary acylcarnitines are altered in human kidney cancer. Int. J. Cancer 2012, 130, 2791–2800. [Google Scholar] [CrossRef]
- Chughtai, K.; Jiang, L.; Greenwood, T.R.; Glunde, K.; Heeren, R.M. Mass spectrometry images acylcarnitines, phosphatidylcholines, and sphingomyelin in MDA-MB-231 breast tumor models. J. Lipid Res. 2013, 54, 333–344. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Samudio, I.; Fiegl, M.; McQueen, T.; Clise-Dwyer, K.; Andreeff, M. The Warburg effect in leukemia-stroma cocultures is mediated by mitochondrial uncoupling associated with uncoupling protein 2 activation. Cancer Res. 2008, 68, 5198–5205. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-L.; Wang, J.-H.; Zhao, A.-H.; Xu, X.; Wang, Y.-H.; Chen, T.-L.; Li, J.-M.; Mi, J.-Q.; Zhu, Y.-M.; Liu, Y.-F. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood 2014, 124, 1645–1654. [Google Scholar] [CrossRef] [PubMed]
- Osowska, S.; Duchemann, T.; Walrand, S.; Paillard, A.; Boirie, Y.; Cynober, L.; Moinard, C. Citrulline modulates muscle protein metabolism in old malnourished rats. Am. J. Physiol.-Endocrinol. Metab. 2006, 291, E582–E586. [Google Scholar] [CrossRef] [PubMed]
- Dowsett, L.; Higgins, E.; Alanazi, S.; Alshuwayer, N.A.; Leiper, F.C.; Leiper, J. ADMA: A key player in the relationship between vascular dysfunction and inflammation in atherosclerosis. J. Clin. Med. 2020, 9, 3026. [Google Scholar] [CrossRef]
| 8 weeks pre-partum | ||||||
| Metabolite, μM1 | CON (SD) | LEU (SD) | P-value | FC | log2(FC)2 | LEU / CON |
| C0 | 3.43 (1.01) | 3.83 (0.89) | 0.361 | 1.11 | 0.16 | UP |
| C2 | 1.60 (0.42) | 1.61 (0.51) | 0.868 | 1.00 | 0.01 | UP |
| C3 | 0.21 (0.05) | 0.19 (0.06) | 0.229 | 0.89 | -0.16 | DOWN |
| C3:1 | 0.03 (0.01) | 0.03 (0.01) | 0.245 | 0.90 | -0.15 | DOWN |
| C3-OH | 0.02 (0.00) | 0.02 (0.00) | 0.245 | 1.08 | 0.11 | UP |
| C4 | 0.08 (0.02) | 0.09 (0.02) | 0.068 | 1.16 | 0.21 | UP |
| C4:1 | 0.02 (0.00) | 0.02 (0.00) | 0.158 | 0.89 | -0.17 | DOWN |
| C4-OH (C3-DC) | 0.03 (0.01) | 0.03 (0.01) | 0.147 | 0.87 | -0.21 | DOWN |
| C5 | 0.07 (0.01) | 0.06 (0.01) | 0.590 | 0.94 | -0.09 | DOWN |
| C5:1 | 0.02 (0.00) | 0.01 (0.00) | 0.171 | 0.88 | -0.18 | DOWN |
| C5:1-DC (C6-OH) | 0.02 (0.00) | 0.02 (0.00) | 0.263 | 0.90 | -0.16 | DOWN |
| C5-DC | 0.01 (0.00) | 0.01 (0.00) | 0.002 | 0.76 | -0.39 | DOWN |
| C5-M-DC | 0.02 (0.00) | 0.02 (0.00) | 0.184 | 0.93 | -0.11 | DOWN |
| C5-OH (C3-DC-M) | 0.07 (0.02) | 0.06 (0.02) | 0.199 | 0.84 | -0.25 | DOWN |
| C6 | 0.05 (0.02) | 0.04 (0.01) | 0.135 | 0.83 | -0.27 | DOWN |
| C6:1 | 0.03 (0.01) | 0.03 (0.01) | 0.010 | 0.83 | -0.27 | DOWN |
| C8 | 0.01 (0.00) | 0.01 (0.01) | 0.507 | 1.04 | 0.06 | UP |
| C9 | 0.01 (0.00) | 0.01 (0.00) | 0.106 | 0.90 | -0.16 | DOWN |
| C10 | 0.05 (0.01) | 0.04 (0.01) | 0.046 | 0.81 | -0.31 | DOWN |
| C10:1 | 0.09 (0.02) | 0.07 (0.03) | 0.023 | 0.75 | -0.42 | DOWN |
| C10:2 | 0.02 (0.01) | 0.02 (0.01) | 0.046 | 0.79 | -0.34 | DOWN |
| C12 | 0.02 (0.00) | 0.02 (0.01) | 0.868 | 1.05 | 0.07 | UP |
| C12:1 | 0.05 (0.01) | 0.04 (0.01) | 0.010 | 0.75 | -0.41 | DOWN |
| C12-DC | 0.02 (0.00) | 0.01 (0.01) | 0.062 | 0.76 | -0.40 | DOWN |
| C14 | 0.02 (0.01) | 0.02 (0.01) | 0.534 | 0.96 | -0.06 | DOWN |
| C14:1 | 0.06 (0.03) | 0.05 (0.02) | 0.431 | 0.86 | -0.22 | DOWN |
| C14:1-OH | 0.01 (0.00) | 0.01 (0.00) | 0.068 | 0.85 | -0.23 | DOWN |
| C14:2 | 0.01 (0.00) | 0.01 (0.00) | 0.455 | 0.92 | -0.12 | DOWN |
| C14:2-OH | 0.01 (0.00) | 0.01 (0.00) | 0.125 | 0.87 | -0.21 | DOWN |
| C16 | 0.02 (0.00) | 0.02 (0.01) | 0.006 | 1.44 | 0.52 | UP |
| C16:1 | 0.02 (0.00) | 0.02 (0.00) | 0.171 | 0.96 | -0.07 | DOWN |
| C16:1-OH | 0.01 (0.00) | 0.01 (0.00) | 0.046 | 0.88 | -0.18 | DOWN |
| C16:2 | 0.01 (0.00) | 0.01 (0.00) | 0.901 | 1.03 | 0.04 | UP |
| C16:2-OH | 0.01 (0.00) | 0.01 (0.00) | 0.034 | 0.82 | -0.29 | DOWN |
| C16-OH | 0.01 (0.00) | 0.01 (0.00) | 0.772 | 0.98 | -0.03 | DOWN |
| C18 | 0.03 (0.01) | 0.04 (0.01) | 0.740 | 1.02 | 0.03 | UP |
| C18:1 | 0.01 (0.00) | 0.02 (0.01) | 0.361 | 1.15 | 0.20 | UP |
| C18:1-OH | 0.01 (0.00) | 0.01 (0.00) | 0.213 | 1.10 | 0.14 | UP |
| C18:2 | 0.01 (0.00) | 0.01 (0.00) | 0.772 | 0.97 | -0.04 | DOWN |
| lyso PC a C14:0 | 1.43 (0.30) | 1.16 (0.30) | 0.038 | 0.81 | -0.31 | DOWN |
| lyso PC a C16:0 | 32.13 (6.27) | 26.34 (9.28) | 0.074 | 0.82 | -0.29 | DOWN |
| lyso PC a C16:1 | 1.89 (0.41) | 1.46 (0.60) | 0.046 | 0.77 | -0.37 | DOWN |
| lyso PC a C17:0 | 1.84 (0.48) | 1.47 (0.63) | 0.125 | 0.80 | -0.32 | DOWN |
| lyso PC a C18:0 | 22.09 (5.35) | 20.06 (6.41) | 0.507 | 0.91 | -0.14 | DOWN |
| lyso PC a C18:1 | 19.97 (4.15) | 14.76 (6.14) | 0.014 | 0.74 | -0.44 | DOWN |
| lyso PC a C18:2 | 45.24 (10.62) | 31.89 (15.21) | 0.014 | 0.70 | -0.50 | DOWN |
| lyso PC a C20:3 | 4.07 (1.07) | 3.95 (1.45) | 1.000 | 0.97 | -0.04 | DOWN |
| lyso PC a C20:4 | 2.66 (0.70) | 2.02 (0.71) | 0.034 | 0.76 | -0.40 | DOWN |
| lyso PC a C24:0 | 0.11 (0.03) | 0.10 (0.03) | 0.245 | 0.89 | -0.17 | DOWN |
| lyso PC a C26:0 | 0.20 (0.09) | 0.15 (0.06) | 0.263 | 0.74 | -0.43 | DOWN |
| lyso PC a C26:1 | 0.07 (0.02) | 0.05 (0.02) | 0.013 | 0.72 | -0.46 | DOWN |
| lyso PC a C28:0 | 0.41 (0.09) | 0.28 (0.10) | 0.001 | 0.68 | -0.57 | DOWN |
| lyso PC a C28:1 | 0.56 (0.12) | 0.42 (0.19) | 0.009 | 0.76 | -0.40 | DOWN |
| PC aa 32:2 | 15.55 (3.35) | 14.41 (5.02) | 0.384 | 0.93 | -0.11 | DOWN |
| PC aa 36:0 | 26.57 (4.35) | 22.99 (7.49) | 0.171 | 0.87 | -0.21 | DOWN |
| PC aa 36:6 | 4.09 (0.95) | 4.38 (1.52) | 0.740 | 1.07 | 0.10 | UP |
| PC aa 38:0 | 4.08 (0.68) | 3.33 (1.11) | 0.042 | 0.82 | -0.29 | DOWN |
| PC aa 38:6 | 4.96 (0.76) | 4.84 (1.53) | 0.678 | 0.98 | -0.04 | DOWN |
| PC aa 40:1 | 0.50 (0.08) | 0.47 (0.14) | 0.772 | 0.93 | -0.11 | DOWN |
| PC aa 40:2 | 2.05 (0.35) | 1.62 (0.59) | 0.020 | 0.79 | -0.34 | DOWN |
| PC aa 40:6 | 2.83 (0.51) | 3.01 (1.05) | 0.934 | 1.06 | 0.09 | UP |
| PC ae 36:0 | 4.29 (0.77) | 3.76 (1.14) | 0.171 | 0.88 | -0.19 | DOWN |
| PC ae 40:6 | 1.57 (0.25) | 1.51 (0.43) | 0.619 | 0.96 | -0.06 | DOWN |
| SM (OH) 14:1 | 14.95 (2.74) | 12.29 (3.58) | 0.042 | 0.82 | -0.28 | DOWN |
| SM 16:0 | 169.89 (28.24) | 152.9 (45.5) | 0.135 | 0.90 | -0.15 | DOWN |
| SM 16:1 | 18.93 (3.07) | 16.25 (4.25) | 0.042 | 0.86 | -0.22 | DOWN |
| SM (OH) 16:1 | 18.14 (3.01) | 14.60 (4.70) | 0.006 | 0.81 | -0.31 | DOWN |
| SM 18:0 | 28.43 (4.66) | 23.32 (6.97) | 0.009 | 0.82 | -0.29 | DOWN |
| SM 18:1 | 30.00 (5.63) | 24.82 (10.86) | 0.014 | 0.83 | -0.27 | DOWN |
| SM 20:2 | 3.62 (0.70) | 3.25 (1.54) | 0.147 | 0.90 | -0.15 | DOWN |
| SM (OH) 22:1 | 31.52 (6.47) | 26.79 (8.24) | 0.089 | 0.85 | -0.23 | DOWN |
| SM (OH) 22:2 | 15.11 (2.71) | 13.14 (3.41) | 0.062 | 0.87 | -0.20 | DOWN |
| SM (OH) 24:1 | 3.38 (0.55) | 2.98 (0.89) | 0.229 | 0.88 | -0.18 | DOWN |
| Glucose | 3,442.97 (281.65) | 3,909.23 (740.35) | 0.062 | 1.14 | 0.18 | UP |
| Alanine | 285.20 (56.09) | 256.20 (57.64) | 0.106 | 0.90 | -0.15 | DOWN |
| Arginine | 124.89 (25.88) | 148.99 (32.75) | 0.042 | 1.19 | 0.25 | UP |
| Asparagine | 36.41 (9.17) | 32.33 (8.96) | 0.213 | 0.89 | -0.17 | DOWN |
| Aspartic acid | 7.86 (2.19) | 11.48 (3.08) | 0.002 | 1.46 | 0.55 | UP |
| Citrulline | 83.22 (22.63) | 78.83 (11.98) | 0.983 | 0.95 | -0.08 | DOWN |
| Glutamine | 276.87 (43.57) | 281.73 (42.49) | 0.836 | 1.02 | 0.03 | UP |
| Glutamic acid | 64.47 (9.87) | 69.47 (17.03) | 0.361 | 1.08 | 0.11 | UP |
| Glycine | 389.07 (64.48) | 328.6 (64.44) | 0.010 | 0.84 | -0.24 | DOWN |
| Histidine | 54.22 (10.86) | 63.29 (13.31) | 0.106 | 1.17 | 0.22 | UP |
| Isoleucine | 117.41 (27.96) | 122.04 (26.49) | 0.455 | 1.04 | 0.06 | UP |
| Leucine | 181.07 (45.40) | 197.33 (59.44) | 0.384 | 1.09 | 0.12 | UP |
| Lysine | 73.73 (20.73) | 86.87 (27.54) | 0.152 | 1.18 | 0.24 | UP |
| Methionine | 22.79 (5.85) | 24.58 (5.06) | 0.431 | 1.08 | 0.11 | UP |
| Ornithine | 47.94 (12.80) | 55.15 (16.22) | 0.221 | 1.15 | 0.20 | UP |
| Phenylalanine | 49.81 (10.19) | 63.54 (17.60) | 0.025 | 1.28 | 0.35 | UP |
| Proline | 113.59 (22.77) | 101.56 (25.07) | 0.097 | 0.89 | -0.16 | DOWN |
| Serine | 90.77 (21.55) | 79.79 (17.49) | 0.130 | 0.88 | -0.19 | DOWN |
| Threonine | 100.71 (27.61) | 97.50 (28.56) | 0.885 | 0.97 | -0.05 | DOWN |
| Tryptophan | 49.03 (8.15) | 51.68 (11.87) | 0.520 | 1.05 | 0.08 | UP |
| Tyrosine | 70.37 (18.02) | 68.43 (19.45) | 0.709 | 0.97 | -0.04 | DOWN |
| Valine | 205.47 (59.31) | 239.73 (60.86) | 0.110 | 1.17 | 0.22 | UP |
| Acetyl-ornithine | 4.01 (1.88) | 3.58 (1.08) | 0.967 | 0.89 | -0.16 | DOWN |
| alpha-Aminoadipic acid | 1.95 (0.51) | 2.11 (1.24) | 0.868 | 1.08 | 0.11 | UP |
| alpha-Ketoglutaric acid | 22.82 (10.13) | 22.97 (7.40) | 0.590 | 1.01 | 0.01 | UP |
| ADMA | 0.65 (0.11) | 0.75 (0.17) | 0.085 | 1.14 | 0.19 | UP |
| beta-Hydroxybutyric acid | 909.13 (353.55) | 721.00 (305.24) | 0.135 | 0.79 | -0.33 | DOWN |
| Betaine | 82.52 (38.26) | 120.57 (67.06) | 0.071 | 1.46 | 0.55 | UP |
| Butyric acid | 18.17 (8.80) | 12.77 (8.58) | 0.101 | 0.70 | -0.51 | DOWN |
| Carnosine | 21.48 (8.82) | 15.45 (5.30) | 0.054 | 0.72 | -0.48 | DOWN |
| Choline | 10.40 (3.21) | 14.01 (3.30) | 0.007 | 1.35 | 0.43 | UP |
| Citric acid | 282.40 (70.10) | 227.92 (90.71) | 0.089 | 0.81 | -0.31 | DOWN |
| Creatine | 248.80 (28.53) | 235.20 (39.61) | 0.319 | 0.95 | -0.08 | DOWN |
| Creatinine | 75.09 (11.60) | 74.33 (11.64) | 0.772 | 0.99 | -0.01 | DOWN |
| Fumaric acid | 1.92 (1.02) | 1.50 (0.93) | 0.152 | 0.78 | -0.36 | DOWN |
| Hippuric acid | 65.31 (13.66) | 65.42 (18.20) | 0.934 | 1.00 | 0.00 | --- |
| Indole acetic acid | 0.59 (0.25) | 0.76 (0.50) | 0.481 | 1.29 | 0.36 | UP |
| Isobutyric acid | 4.82 (1.71) | 6.91 (4.21) | 0.147 | 1.43 | 0.52 | UP |
| Kynurenine | 9.06 (2.72) | 8.60 (4.12) | 0.648 | 0.95 | -0.07 | DOWN |
| Lactic acid | 1,489.93 (729.91) | 1,514.73 (720.62) | 0.917 | 1.02 | 0.02 | UP |
| Methionine-sulfoxide | 2.47 (0.62) | 2.14 (0.71) | 0.237 | 0.87 | -0.21 | DOWN |
| Methylhistidine | 9.37 (2.63) | 10.02 (2.49) | 0.468 | 1.07 | 0.10 | UP |
| Methylmalonic acid | 0.60 (0.13) | 0.64 (0.47) | 0.263 | 1.07 | 0.09 | UP |
| Propionic acid | 35.89 (18.95) | 29.61 (15.44) | 0.431 | 0.83 | -0.28 | DOWN |
| Pyruvic acid | 57.17 (13.86) | 67.69 (16.98) | 0.074 | 1.18 | 0.24 | UP |
| Serotonin | 7.25 (5.04) | 8.08 (6.42) | 0.934 | 1.12 | 0.16 | UP |
| Spermidine | 0.29 (0.12) | 0.29 (0.14) | 0.836 | 1.02 | 0.02 | UP |
| Succinic acid | 1.48 (0.31) | 1.37 (0.43) | 0.130 | 0.93 | -0.11 | DOWN |
| Taurine | 75.57 (20.50) | 73.01 (14.75) | 0.803 | 0.97 | -0.05 | DOWN |
| TDMA | 1.92 (0.41) | 1.79 (0.36) | 0.836 | 0.93 | -0.10 | DOWN |
| trans-Hydroxyproline | 12.57 (2.65) | 10.83 (2.49) | 0.018 | 0.86 | -0.22 | DOWN |
| Trimethylamine N-oxide | 47.05 (33.10) | 46.28 (44.15) | 0.619 | 0.98 | -0.02 | DOWN |
| Uric acid | 37.16 (12.94) | 46.48 (24.56) | 0.158 | 1.25 | 0.32 | UP |
| 4 weeks pre-partum | |||||||
| Metabolite, μM1 | CON (SD) | LEU (SD) | P-value | FC | log2(FC)2 | LEU / CON | |
| C0 | 4.76 (1.62) | 4.18 (1.20) | 0.431 | 0.88 | -0.19 | DOWN | |
| C2 | 2.01 (0.57) | 1.93 (0.46) | 0.709 | 0.96 | -0.06 | DOWN | |
| C3 | 0.19 (0.05) | 0.18 (0.04) | 0.590 | 0.92 | -0.11 | DOWN | |
| C3:1 | 0.02 (0.00) | 0.02 (0.01) | 0.340 | 0.93 | -0.11 | DOWN | |
| C3-OH | 0.02 (0.01) | 0.02 (0.00) | 0.002 | 0.69 | -0.53 | DOWN | |
| C4 | 0.10 (0.02) | 0.10 (0.03) | 0.648 | 1.04 | 0.06 | UP | |
| C4:1 | 0.02 (0.00) | 0.01 (0.00) | 0.678 | 0.96 | -0.06 | DOWN | |
| C4-OH (C3-DC) | 0.03 (0.01) | 0.03 (0.01) | 0.561 | 0.98 | -0.03 | DOWN | |
| C5 | 0.08 (0.01) | 0.07 (0.02) | 0.184 | 0.89 | -0.17 | DOWN | |
| C5:1 | 0.01 (0.00) | 0.01 (0.00) | 0.020 | 0.81 | -0.31 | DOWN | |
| C5:1-DC (C6-OH) | 0.01 (0.00) | 0.01 (0.00) | 0.147 | 1.13 | 0.17 | UP | |
| C5-DC | 0.01 (0.00) | 0.01 (0.00) | 0.407 | 0.94 | -0.09 | DOWN | |
| C5-M-DC | 0.02 (0.00) | 0.02 (0.00) | 0.005 | 1.15 | 0.20 | UP | |
| C5-OH (C3-DC-M) | 0.06 (0.03) | 0.08 (0.02) | 0.038 | 1.42 | 0.51 | UP | |
| C6 | 0.03 (0.01) | 0.05 (0.04) | 0.081 | 1.58 | 0.66 | UP | |
| C6:1 | 0.02 (0.01) | 0.03 (0.01) | 0.619 | 1.12 | 0.16 | UP | |
| C8 | 0.01 (0.00) | 0.01 (0.01) | 0.590 | 1.08 | 0.11 | UP | |
| C9 | 0.01 (0.00) | 0.01 (0.00) | 0.147 | 0.92 | -0.12 | DOWN | |
| C10 | 0.04 (0.01) | 0.03 (0.01) | 0.534 | 0.9 | -0.15 | DOWN | |
| C10:1 | 0.11 (0.06) | 0.08 (0.03) | 0.300 | 0.68 | -0.56 | DOWN | |
| C10:2 | 0.02 (0.01) | 0.02 (0.00) | 0.901 | 0.95 | -0.08 | DOWN | |
| C12 | 0.02 (0.00) | 0.03 (0.01) | 0.590 | 1.14 | 0.19 | UP | |
| C12:1 | 0.05 (0.03) | 0.04 (0.01) | 0.281 | 0.73 | -0.46 | DOWN | |
| C12-DC | 0.01 (0.00) | 0.01 (0.00) | 0.281 | 1.09 | 0.13 | UP | |
| C14 | 0.01 (0.00) | 0.01 (0.01) | 0.407 | 1.08 | 0.11 | UP | |
| C14:1 | 0.05 (0.02) | 0.04 (0.01) | 0.068 | 0.83 | -0.26 | DOWN | |
| C14:1-OH | 0.01 (0.00) | 0.01 (0.00) | 0.431 | 1.08 | 0.11 | UP | |
| C14:2 | 0.01 (0.00) | 0.01 (0.00) | 0.648 | 1.07 | 0.10 | UP | |
| C14:2-OH | 0.01 (0.00) | 0.01 (0.00) | 1.000 | 0.96 | -0.06 | DOWN | |
| C16 | 0.02 (0.00) | 0.03 (0.01) | 0.709 | 1.15 | 0.20 | UP | |
| C16:1 | 0.02 (0.00) | 0.02 (0.00) | 0.213 | 0.97 | -0.04 | DOWN | |
| C16:1-OH | 0.01 (0.00) | 0.01 (0.00) | 0.868 | 0.98 | -0.03 | DOWN | |
| C16:2 | 0.01 (0.00) | 0.01 (0.00) | 0.507 | 0.96 | -0.06 | DOWN | |
| C16:2-OH | 0.01 (0.00) | 0.01 (0.00) | 0.135 | 0.87 | -0.21 | DOWN | |
| C16-OH | 0.01 (0.00) | 0.01 (0.00) | 0.056 | 0.85 | -0.23 | DOWN | |
| C18 | 0.03 (0.01) | 0.03 (0.01) | 0.967 | 1.08 | 0.11 | UP | |
| C18:1 | 0.02 (0.00) | 0.02 (0.01) | 0.320 | 1.23 | 0.30 | UP | |
| C18:1-OH | 0.01 (0.00) | 0.01 (0.00) | 0.184 | 0.86 | -0.22 | DOWN | |
| C18:2 | 0.01 (0.00) | 0.01 (0.00) | 0.934 | 0.98 | -0.03 | DOWN | |
| lyso PC a C14:0 | 0.84 (0.25) | 0.76 (0.21) | 0.320 | 0.91 | -0.14 | DOWN | |
| lyso PC a C16:0 | 17.78 (4.36) | 17.35 (4.67) | 0.868 | 0.98 | -0.03 | DOWN | |
| lyso PC a C16:1 | 1.13 (0.37) | 1.16 (0.42) | 0.481 | 1.03 | 0.04 | UP | |
| lyso PC a C17:0 | 1.29 (0.27) | 1.29 (0.48) | 0.868 | 1.00 | 0.01 | UP | |
| lyso PC a C18:0 | 15.89 (3.24) | 13.33 (2.88) | 0.042 | 0.84 | -0.25 | DOWN | |
| lyso PC a C18:1 | 11.18 (3.40) | 10.75 (3.63) | 0.967 | 0.96 | -0.06 | DOWN | |
| lyso PC a C18:2 | 19.96 (5.95) | 18.42 (6.67) | 0.534 | 0.92 | -0.12 | DOWN | |
| lyso PC a C20:3 | 3.32 (1.02) | 2.79 (0.81) | 0.135 | 0.84 | -0.25 | DOWN | |
| lyso PC a C20:4 | 1.82 (0.54) | 1.77 (0.65) | 0.868 | 0.97 | -0.04 | DOWN | |
| lyso PC a C24:0 | 0.11 (0.03) | 0.10 (0.01) | 0.507 | 0.93 | -0.10 | DOWN | |
| lyso PC a C26:0 | 0.08 (0.03) | 0.07 (0.02) | 0.229 | 0.83 | -0.27 | DOWN | |
| lyso PC a C26:1 | 0.04 (0.01) | 0.04 (0.01) | 0.803 | 0.98 | -0.03 | DOWN | |
| lyso PC a C28:0 | 0.23 (0.07) | 0.21 (0.08) | 0.245 | 0.90 | -0.15 | DOWN | |
| lyso PC a C28:1 | 0.31 (0.11) | 0.27 (0.13) | 0.171 | 0.86 | -0.22 | DOWN | |
| PC aa 32:2 | 11.85 (2.90) | 9.83 (3.24) | 0.074 | 0.83 | -0.27 | DOWN | |
| PC aa 36:0 | 14.48 (2.02) | 11.94 (4.01) | 0.028 | 0.82 | -0.28 | DOWN | |
| PC aa 36:6 | 3.59 (0.93) | 2.98 (1.02) | 0.074 | 0.83 | -0.27 | DOWN | |
| PC aa 38:0 | 1.93 (0.30) | 1.39 (0.58) | 0.005 | 0.72 | -0.47 | DOWN | |
| PC aa 38:6 | 3.45 (0.68) | 2.75 (0.63) | 0.010 | 0.80 | -0.32 | DOWN | |
| PC aa 40:1 | 0.40 (0.08) | 0.33 (0.09) | 0.046 | 0.83 | -0.26 | DOWN | |
| PC aa 40:2 | 0.99 (0.18) | 0.71 (0.25) | 0.004 | 0.72 | -0.48 | DOWN | |
| PC aa 40:6 | 2.54 (0.43) | 2.21 (0.57) | 0.081 | 0.87 | -0.20 | DOWN | |
| PC ae 36:0 | 3.38 (0.68) | 2.71 (0.91) | 0.034 | 0.8 | -0.32 | DOWN | |
| PC ae 40:6 | 1.15 (0.18) | 0.95 (0.23) | 0.011 | 0.83 | -0.28 | DOWN | |
| SM (OH) 14:1 | 9.30 (2.06) | 7.78 (2.10) | 0.068 | 0.84 | -0.26 | DOWN | |
| SM 16:0 | 101.36 (25.24) | 89.70 (21.76) | 0.171 | 0.88 | -0.18 | DOWN | |
| SM 16:1 | 11.21 (2.63) | 9.70 (2.44) | 0.135 | 0.87 | -0.21 | DOWN | |
| SM (OH) 16:1 | 9.92 (2.45) | 8.86 (2.55) | 0.300 | 0.89 | -0.16 | DOWN | |
| SM 18:0 | 17.23 (4.27) | 15.98 (3.88) | 0.340 | 0.93 | -0.11 | DOWN | |
| SM 18:1 | 16.60 (5.51) | 15.45 (4.87) | 0.740 | 0.93 | -0.10 | DOWN | |
| SM 20:2 | 2.41 (0.95) | 2.18 (0.79) | 0.678 | 0.9 | -0.15 | DOWN | |
| SM (OH) 22:1 | 18.13 (4.72) | 17.42 (4.74) | 0.648 | 0.96 | -0.06 | DOWN | |
| SM (OH) 22:2 | 10.01 (2.53) | 9.33 (2.50) | 0.361 | 0.93 | -0.10 | DOWN | |
| SM (OH) 24:1 | 2.27 (0.63) | 2.32 (0.56) | 0.772 | 1.02 | 0.03 | UP | |
| Glucose | 3,900.8 (423.3) | 3,855.92 (376.37) | 0.678 | 0.99 | -0.02 | DOWN | |
| Alanine | 2.16 (1.32) | 1.99 (0.67) | 0.868 | 0.92 | -0.12 | DOWN | |
| Arginine | 30.46 (4.57) | 29.38 (7.37) | 0.590 | 0.96 | -0.05 | DOWN | |
| Asparagine | 8.15 (2.88) | 9.89 (2.68) | 0.130 | 1.21 | 0.28 | UP | |
| Aspartic acid | 1.07 (0.19) | 1.02 (0.13) | 0.220 | 0.95 | -0.07 | DOWN | |
| Citrulline | 85.98 (16.86) | 85.55 (14.21) | 0.836 | 0.99 | -0.01 | DOWN | |
| Glutamine | 330.33 (43.70) | 317.20 (46.41) | 0.350 | 0.96 | -0.06 | DOWN | |
| Glutamic acid | 68.43 (21.47) | 69.57 (19.99) | 0.885 | 1.02 | 0.02 | UP | |
| Glycine | 262.47 (31.35) | 236.60 (26.17) | 0.026 | 0.90 | -0.15 | DOWN | |
| Histidine | 71.49 (9.49) | 68.01 (10.49) | 0.395 | 0.95 | -0.07 | DOWN | |
| Isoleucine | 145.27 (17.73) | 149.87 (24.80) | 0.618 | 1.03 | 0.04 | UP | |
| Leucine | 232.53 (67.47) | 244.47 (46.21) | 0.213 | 1.05 | 0.07 | UP | |
| Lysine | 96.77 (30.14) | 103.05 (21.40) | 0.290 | 1.06 | 0.09 | UP | |
| Methionine | 29.46 (5.08) | 27.95 (4.84) | 0.520 | 0.95 | -0.08 | DOWN | |
| Ornithine | 63.85 (11.38) | 60.23 (10.44) | 0.395 | 0.94 | -0.08 | DOWN | |
| Phenylalanine | 63.49 (9.68) | 66.11 (7.48) | 0.178 | 1.04 | 0.06 | UP | |
| Proline | 97.81 (14.53) | 81.47 (16.86) | 0.012 | 0.83 | -0.26 | DOWN | |
| Serine | 74.99 (14.60) | 70.39 (13.21) | 0.633 | 0.94 | -0.09 | DOWN | |
| Threonine | 2.22 (0.33) | 2.47 (0.40) | 0.054 | 1.11 | 0.16 | UP | |
| Tryptophan | 43.63 (7.99) | 41.67 (6.73) | 0.967 | 0.96 | -0.07 | DOWN | |
| Tyrosine | 64.95 (15.17) | 64.10 (12.77) | 0.934 | 0.99 | -0.02 | DOWN | |
| Valine | 294.07 (48.43) | 262.80 (52.10) | 0.184 | 0.89 | -0.16 | DOWN | |
| Acetyl-ornithine | 3.86 (1.22) | 3.28 (1.59) | 0.272 | 0.85 | -0.24 | DOWN | |
| alpha-Aminoadipic acid | 16.94 (5.38) | 19.44 (3.49) | 0.068 | 1.15 | 0.20 | UP | |
| alpha-Ketoglutaric acid | 148.47 (25.11) | 136.33 (26.66) | 0.089 | 0.92 | -0.12 | DOWN | |
| ADMA | 240.07 (32.79) | 212.73 (41.04) | 0.026 | 0.89 | -0.17 | DOWN | |
| beta-Hydroxybutyric acid | 676.20 (261.66) | 520.80 (147.68) | 0.078 | 0.77 | -0.38 | DOWN | |
| Betaine | 164.53 (37.37) | 150.69 (46.30) | 0.494 | 0.92 | -0.13 | DOWN | |
| Butyric acid | 7.26 (2.43) | 5.35 (2.73) | 0.065 | 0.74 | -0.44 | DOWN | |
| Carnosine | 13.01 (4.63) | 9.26 (4.83) | 0.033 | 0.71 | -0.49 | DOWN | |
| Choline | 11.75 (2.89) | 12.53 (5.37) | 0.724 | 1.07 | 0.09 | UP | |
| Citric acid | 292.67 (110.61) | 227.25 (82.03) | 0.068 | 0.78 | -0.36 | DOWN | |
| Creatine | 242.67 (33.07) | 240.07 (33.93) | 0.884 | 0.99 | -0.02 | DOWN | |
| Creatinine | 75.09 (11.60) | 74.33 (11.64) | 0.772 | 0.99 | -0.01 | DOWN | |
| Fumaric acid | 1.07 (0.24) | 1.15 (0.22) | 0.372 | 1.07 | 0.10 | UP | |
| Hippuric acid | 71.57 (15.24) | 66.83 (15.73) | 0.351 | 0.93 | -0.10 | DOWN | |
| Indole acetic acid | 0.40 (0.16) | 0.36 (0.18) | 0.35 | 0.92 | -0.13 | DOWN | |
| Isobutyric acid | 5.22 (1.62) | 4.65 (1.48) | 0.351 | 0.89 | -0.17 | DOWN | |
| Kynurenine | 6.66 (2.40) | 7.15 (3.78) | 0.633 | 1.07 | 0.10 | UP | |
| Lactic acid | 1,420.53 (680.05) | 1,362.27 (819.95) | 0.361 | 0.96 | -0.06 | DOWN | |
| Methionine-sulfoxide | 2.82 (0.81) | 1.79 (0.58) | 0.001 | 0.64 | -0.65 | DOWN | |
| Methylhistidine | 13.87 (4.22) | 12.91 (3.34) | 0.756 | 0.93 | -0.10 | DOWN | |
| Methylmalonic acid | 0.35 (0.13) | 0.37 (0.18) | 0.836 | 1.07 | 0.09 | UP | |
| Propionic acid | 19.64 (7.54) | 16.83 (7.92) | 0.418 | 0.86 | -0.22 | DOWN | |
| Pyruvic acid | 67.15 (17.04) | 69.96 (23.32) | 0.917 | 1.04 | 0.06 | UP | |
| Serotonin | 7.94 (9.72) | 4.88 (6.11) | 0.290 | 0.61 | -0.70 | DOWN | |
| Spermidine | 0.31 (0.17) | 0.22 (0.10) | 0.165 | 0.70 | -0.51 | DOWN | |
| Succinic acid | 1.15 (0.27) | 1.07 (0.24) | 0.547 | 0.93 | -0.11 | DOWN | |
| Taurine | 71.39 (23.64) | 67.73 (21.07) | 0.756 | 0.95 | -0.08 | DOWN | |
| TDMA | 98.41 (24.02) | 89.70 (16.86) | 0.885 | 0.91 | -0.13 | DOWN | |
| trans-Hydroxyproline | 15.41 (2.86) | 13.13 (3.25) | 0.085 | 0.85 | -0.23 | DOWN | |
| Trimethylamine N-oxide | 23.65 (24.39) | 13.34 (10.02) | 0.229 | 0.56 | -0.83 | DOWN | |
| Uric acid | 37.38 (28.74) | 40.21 (27.23) | 0.372 | 1.08 | 0.11 | UP | |
| 8 weeks pre-partum | |||||||||
| Metabolite, μM1 | CON (SD) | LEU (SD) | P-value | FC | log2(FC)2 | LEU / CON | |||
| C0 | 1.12 (0.61) | 1.26 (0.55) | 0.561 | 1.12 | 0.17 | UP | |||
| C2 | 0.34 (0.10) | 0.45 (0.15) | 0.081 | 1.33 | 0.41 | UP | |||
| C3 | 0.03 (0.01) | 0.03 (0.02) | 0.534 | 1.11 | 0.15 | UP | |||
| C3:1 | 0.02 (0.01) | 0.02 (0.01) | 0.407 | 1.17 | 0.23 | UP | |||
| C3-OH | 0.07 (0.02) | 0.07 (0.02) | 0.431 | 1.05 | 0.07 | UP | |||
| C4 | 0.19 (0.10) | 0.28 (0.14) | 0.023 | 1.49 | 0.58 | UP | |||
| C4:1 | 0.06 (0.02) | 0.07 (0.03) | 0.281 | 1.16 | 0.22 | UP | |||
| C4-OH (C3-DC) | 0.07 (0.02) | 0.07 (0.02) | 0.648 | 0.95 | -0.08 | DOWN | |||
| C5 | 0.12 (0.05) | 0.11 (0.05) | 0.836 | 0.93 | -0.10 | DOWN | |||
| C5:1 | 0.14 (0.06) | 0.14 (0.05) | 0.868 | 1.03 | 0.04 | UP | |||
| C5:1-DC (C6-OH) | 0.03 (0.01) | 0.04 (0.01) | 1.000 | 1.04 | 0.06 | UP | |||
| C5-DC | 0.03 (0.01) | 0.03 (0.01) | 0.803 | 1.05 | 0.07 | UP | |||
| C5-M-DC | 0.04 (0.01) | 0.05 (0.01) | 0.074 | 1.21 | 0.28 | UP | |||
| C5-OH (C3-DC-M) | 0.08 (0.03) | 0.09 (0.02) | 0.740 | 1.03 | 0.04 | UP | |||
| C6 | 0.08 (0.04) | 0.08 (0.03) | 0.590 | 1.06 | 0.09 | UP | |||
| C6:1 | 0.07 (0.03) | 0.08 (0.02) | 0.455 | 1.11 | 0.14 | UP | |||
| C7-DC | 0.03 (0.02) | 0.03 (0.01) | 0.934 | 0.93 | -0.11 | DOWN | |||
| C8 | 0.03 (0.02) | 0.04 (0.02) | 0.056 | 1.34 | 0.43 | UP | |||
| C9 | 0.10 (0.06) | 0.10 (0.04) | 0.740 | 0.99 | -0.02 | DOWN | |||
| C10 | 0.09 (0.02) | 0.12 (0.03) | 0.004 | 1.31 | 0.39 | UP | |||
| C10:1 | 0.12 (0.04) | 0.17 (0.04) | 0.010 | 1.37 | 0.45 | UP | |||
| C10:2 | 0.04 (0.01) | 0.05 (0.01) | 0.051 | 1.20 | 0.27 | UP | |||
| C12 | 0.07 (0.02) | 0.12 (0.10) | 0.003 | 1.88 | 0.91 | UP | |||
| C12:1 | 0.11 (0.06) | 0.17 (0.07) | 0.018 | 1.47 | 0.55 | UP | |||
| Glucose | 1,244.85 (634.77) | 2,479.37 (1,200.03) | 0.003 | 1.99 | 0.99 | UP | |||
| Alanine | 248.59 (118.99) | 197.04 (97.34) | 0.340 | 0.79 | -0.34 | DOWN | |||
| Arginine | 9.08 (5.12) | 12.74 (8.54) | 0.213 | 1.40 | 0.49 | UP | |||
| Asparagine | 8.76 (4.69) | 11.63 (7.57) | 0.281 | 1.33 | 0.41 | UP | |||
| Aspartic acid | 132.49 (103.51) | 131.56 (72.13) | 0.709 | 0.99 | -0.01 | DOWN | |||
| Citrulline | 4.55 (3.59) | 7.07 (4.38) | 0.046 | 1.55 | 0.63 | UP | |||
| Glutamine | 258.05 (165.24) | 292.15 (177.73) | 0.678 | 1.13 | 0.18 | UP | |||
| Glutamic acid | 103.23 (66.24) | 92.03 (48.27) | 0.709 | 0.89 | -0.17 | DOWN | |||
| Glycine | 351.55 (437.92) | 250.31 (221.53) | 1.000 | 0.71 | -0.49 | DOWN | |||
| Histidine | 77.01 (60.57) | 82.78 (52.01) | 0.561 | 1.07 | 0.10 | UP | |||
| Isoleucine | 12.00 (12.97) | 10.78 (7.31) | 0.534 | 0.90 | -0.16 | DOWN | |||
| Leucine | 6.79 (3.74) | 8.65 (4.06) | 0.120 | 1.27 | 0.35 | UP | |||
| Lysine | 53.07 (34.93) | 60.67 (33.51) | 0.455 | 1.14 | 0.19 | UP | |||
| Methionine | 2.71 (0.68) | 3.13 (0.91) | 0.191 | 1.16 | 0.21 | UP | |||
| Ornithine | 17.11 (10.12) | 19.45 (6.95) | 0.237 | 1.14 | 0.19 | UP | |||
| Phenylalanine | 11.30 (6.90) | 14.64 (11.75) | 0.407 | 1.30 | 0.37 | UP | |||
| Proline | 7.41 (4.52) | 5.67 (2.77) | 0.520 | 0.76 | -0.39 | DOWN | |||
| Serine | 74.59 (46.99) | 84.83 (40.33) | 0.320 | 1.14 | 0.19 | UP | |||
| Threonine | 67.11 (39.41) | 68.59 (35.24) | 0.917 | 1.02 | 0.03 | UP | |||
| Tryptophan | 27.26 (16.29) | 25.17 (12.73) | 0.885 | 0.92 | -0.12 | DOWN | |||
| Tyrosine | 21.37 (10.49) | 22.40 (9.46) | 0.678 | 1.05 | 0.07 | UP | |||
| Valine | 10.50 (6.91) | 11.54 (4.60) | 0.320 | 1.10 | 0.14 | UP | |||
| Acetyl-ornithine | 51.89 (34.54) | 56.36 (25.04) | 0.340 | 1.09 | 0.12 | UP | |||
| ADMA | 3.07 (1.87) | 6.03 (4.57) | 0.023 | 1.97 | 0.98 | UP | |||
| alpha-Aminoadipic acid | 107.94 (70.37) | 98.35 (56.85) | 0.772 | 0.91 | -0.13 | DOWN | |||
| alpha-Ketoglutaric acid | 47.65 (124.98) | 53.00 (160.42) | 0.619 | 1.11 | 0.15 | UP | |||
| beta-Hydroxybutyric acid | 664.87 (1,242.58) | 286.33 (525.75) | 0.213 | 0.43 | -1.22 | DOWN | |||
| Betaine | 113.17 (89.01) | 204.88 (164.69) | 0.097 | 1.81 | 0.86 | UP | |||
| Butyric acid | 43.40 (39.29) | 34.30 (38.63) | 0.213 | 0.79 | -0.34 | DOWN | |||
| Carnosine | 16.37 (11.21) | 14.72 (8.95) | 0.561 | 0.90 | -0.15 | DOWN | |||
| Choline | 26.75 (25.65) | 50.75 (63.00) | 0.351 | 1.90 | 0.92 | UP | |||
| Citric acid | 976.90 (1,968.16) | 984.5 (1,735.92) | 0.619 | 1.01 | 0.01 | UP | |||
| Creatine | 3,140.67 (1,603.8) | 4,036.13 (1,691.91) | 0.184 | 1.29 | 0.36 | UP | |||
| Creatinine | 8,130.67 (4,491.35) | 9,936.67 (5,070.74) | 0.300 | 1.22 | 0.29 | UP | |||
| Hippuric acid | 13,707.33 (4,854.71) | 12,136.00 (2,795.60) | 0.480 | 0.89 | -0.18 | DOWN | |||
| Histamine | 0.10 (0.06) | 0.11 (0.06) | 0.633 | 1.07 | 0.10 | UP | |||
| Homovanillic acid | 6.59 (4.19) | 7.93 (5.62) | 0.431 | 1.20 | 0.27 | UP | |||
| Indole acetic acid | 66.79 (54.74) | 66.61 (45.70) | 0.852 | 1.00 | 0.00 | --- | |||
| Isobutyric acid | 6.01 (4.30) | 10.88 (15.96) | 0.619 | 1.81 | 0.86 | UP | |||
| Kynurenine | 1.37 (1.11) | 1.06 (0.63) | 0.709 | 0.77 | -0.37 | DOWN | |||
| Lactic acid | 108.56 (146.26) | 96.51 (73.37) | 0.431 | 0.89 | -0.17 | DOWN | |||
| Methionine-sulfoxide | 2.65 (2.19) | 2.29 (1.53) | 0.967 | 0.86 | -0.21 | DOWN | |||
| Methylhistidine | 224.15 (148.69) | 266.00 (171.06) | 0.431 | 1.19 | 0.25 | UP | |||
| Methylmalonic acid | 23.00 (17.27) | 24.75 (23.39) | 0.885 | 1.08 | 0.11 | UP | |||
| p-Hydroxyhippuric acid | 43.62 (39.85) | 39.65 (28.06) | 0.950 | 0.91 | -0.14 | DOWN | |||
| Putrescine | 0.42 (0.22) | 0.62 (0.57) | 0.852 | 1.47 | 0.55 | UP | |||
| Pyruvic acid | 5.01 (4.48) | 6.21 (4.36) | 0.141 | 1.24 | 0.31 | UP | |||
| Sarcosine | 2.72 (2.26) | 4.14 (3.41) | 0.213 | 1.52 | 0.61 | UP | |||
| Serotonin | 1.43 (0.84) | 1.55 (0.74) | 0.419 | 1.09 | 0.12 | UP | |||
| Spermidine | 0.08 (0.05) | 0.25 (0.57) | 0.237 | 3.09 | 1.63 | UP | |||
| Spermine | 0.08 (0.03) | 0.10 (0.05) | 0.135 | 1.36 | 0.45 | UP | |||
| Succinic acid | 22.30 (23.99) | 26.10 (20.52) | 0.330 | 1.17 | 0.23 | UP | |||
| Taurine | 492.27 (512.53) | 653.27 (522.13) | 0.263 | 1.33 | 0.41 | UP | |||
| TDMA | 20.38 (10.71) | 26.82 (14.60) | 0.229 | 1.32 | 0.40 | UP | |||
| trans-Hydroxyproline | 1.61 (1.53) | 1.35 (0.79) | 0.852 | 0.84 | -0.26 | DOWN | |||
| Trimethylamine N-oxide | 3,960.93 (3,391.24) | 4,052.87 (4,086.49) | 1.000 | 1.02 | 0.03 | UP | |||
| Tyramine | 0.08 (0.08) | 0.14 (0.12) | 0.088 | 1.75 | 0.81 | UP | |||
| Uric acid | 4,122.67 (2,408.00) | 4,300.00 (2,455.60) | 0.772 | 1.04 | 0.06 | UP | |||
| 4 weeks pre-partum | ||||||
| Metabolite, μM1 | CON (SD) | LEU (SD) | P-value | FC | log2(FC)2 | LEU / CON |
| C0 | 1.17 (0.32) | 1.27 (0.49) | 0.561 | 1.09 | 0.12 | UP |
| C2 | 0.64 (0.32) | 0.52 (0.21) | 0.431 | 0.82 | -0.29 | DOWN |
| C3 | 0.06 (0.01) | 0.05 (0.01) | 0.056 | 0.91 | -0.14 | DOWN |
| C3:1 | 0.05 (0.01) | 0.05 (0.02) | 0.648 | 1.00 | -0.01 | DOWN |
| C3-OH | 0.07 (0.02) | 0.07 (0.02) | 1.000 | 1.01 | 0.02 | UP |
| C4 | 0.38 (0.22) | 0.42 (0.19) | 0.534 | 1.11 | 0.14 | UP |
| C4:1 | 0.07 (0.02) | 0.07 (0.02) | 0.619 | 0.99 | -0.02 | DOWN |
| C4-OH (C3-DC) | 0.07 (0.02) | 0.07 (0.02) | 0.901 | 1.00 | 0.00 | --- |
| C5 | 0.15 (0.06) | 0.12 (0.04) | 0.481 | 0.84 | -0.25 | DOWN |
| C5:1 | 0.17 (0.03) | 0.20 (0.13) | 0.709 | 1.20 | 0.26 | UP |
| C5:1-DC (C6-OH) | 0.04 (0.01) | 0.03 (0.01) | 0.534 | 0.97 | -0.05 | DOWN |
| C5-DC | 0.03 (0.01) | 0.03 (0.02) | 0.709 | 1.13 | 0.17 | UP |
| C5-M-DC | 0.05 (0.01) | 0.05 (0.02) | 0.934 | 1.02 | 0.02 | UP |
| C5-OH (C3-DC-M) | 0.11 (0.02) | 0.12 (0.04) | 0.934 | 1.06 | 0.08 | UP |
| C6 | 0.09 (0.02) | 0.09 (0.03) | 0.534 | 0.92 | -0.11 | DOWN |
| C6:1 | 0.08 (0.02) | 0.08 (0.02) | 0.384 | 1.04 | 0.05 | UP |
| C7-DC | 0.03 (0.01) | 0.04 (0.02) | 0.245 | 1.25 | 0.32 | UP |
| C8 | 0.04 (0.01) | 0.04 (0.01) | 0.740 | 0.98 | -0.03 | DOWN |
| C9 | 0.12 (0.04) | 0.13 (0.06) | 0.740 | 1.08 | 0.12 | UP |
| C10 | 0.13 (0.04) | 0.13 (0.05) | 0.901 | 0.98 | -0.02 | DOWN |
| C10:1 | 0.18 (0.04) | 0.20 (0.05) | 0.229 | 1.13 | 0.17 | UP |
| C10:2 | 0.05 (0.02) | 0.05 (0.03) | 0.648 | 1.06 | 0.08 | UP |
| C12 | 0.11 (0.05) | 0.09 (0.06) | 0.320 | 0.89 | -0.17 | DOWN |
| C12:1 | 0.21 (0.1) | 0.20 (0.12) | 0.648 | 0.93 | -0.10 | DOWN |
| Glucose | 1,600.88 (1,218.21) | 1,253.64 (944.4) | 0.431 | 0.78 | -0.35 | DOWN |
| Alanine | 83.23 (25.77) | 89.55 (38.33) | 0.494 | 1.08 | 0.11 | UP |
| Arginine | 11.32 (3.30) | 12.08 (5.55) | 0.663 | 1.07 | 0.09 | UP |
| Asparagine | 9.52 (2.98) | 10.86 (5.54) | 0.395 | 1.14 | 0.19 | UP |
| Aspartic acid | 140.83 (60.83) | 144.25 (73.88) | 0.82 | 1.02 | 0.03 | UP |
| Citrulline | 6.76 (4.16) | 8.41 (6.88) | 0.709 | 1.24 | 0.32 | UP |
| Glutamine | 215.95 (85.79) | 245.27 (136.20) | 0.633 | 1.14 | 0.18 | UP |
| Glutamic acid | 57.76 (16.75) | 62.01 (35.25) | 0.885 | 1.07 | 0.10 | UP |
| Glycine | 73.81 (33.98) | 83.26 (48.58) | 0.431 | 1.13 | 0.17 | UP |
| Histidine | 64.61 (16.23) | 75.67 (42.70) | 0.431 | 1.17 | 0.23 | UP |
| Isoleucine | 6.07 (1.22) | 6.82 (3.08) | 0.330 | 1.12 | 0.17 | UP |
| Leucine | 9.15 (2.51) | 10.43 (4.79) | 0.633 | 1.14 | 0.19 | UP |
| Lysine | 50.96 (11.56) | 58.83 (32.78) | 0.604 | 1.15 | 0.21 | UP |
| Methionine | 3.23 (0.50) | 3.29 (0.86) | 0.836 | 1.02 | 0.03 | UP |
| Ornithine | 16.28 (5.10) | 17.59 (9.46) | 0.756 | 1.08 | 0.11 | UP |
| Phenylalanine | 11.04 (2.70) | 13.21 (7.76) | 0.372 | 1.20 | 0.26 | UP |
| Proline | 3.99 (1.21) | 3.87 (1.79) | 0.772 | 0.97 | -0.04 | DOWN |
| Serine | 67.39 (23.68) | 78.19 (33.86) | 0.384 | 1.16 | 0.21 | UP |
| Threonine | 51.43 (17.40) | 58.97 (37.45) | 0.663 | 1.15 | 0.20 | UP |
| Tryptophan | 19.51 (6.70) | 21.41 (14.62) | 0.756 | 1.10 | 0.13 | UP |
| Tyrosine | 21.25 (5.43) | 23.76 (13.32) | 0.803 | 1.12 | 0.16 | UP |
| Valine | 10.45 (2.59) | 11.64 (4.64) | 0.395 | 1.11 | 0.16 | UP |
| Acetyl-ornithine | 46.31 (14.14) | 62.25 (33.27) | 0.135 | 1.34 | 0.43 | UP |
| ADMA | 6.78 (2.53) | 7.73 (4.06) | 0.507 | 1.14 | 0.19 | UP |
| alpha-Aminoadipic acid | 78.13 (32.65) | 75.09 (28.38) | 0.983 | 0.96 | -0.06 | DOWN |
| alpha-Ketoglutaric acid | 9.66 (12.05) | 52.06 (147.06) | 0.184 | 5.39 | 2.43 | UP |
| beta-Hydroxybutyric acid | 133.04 (185.33) | 83.27 (75.78) | 0.934 | 0.63 | -0.68 | DOWN |
| Betaine | 358.29 (286.02) | 280.29 (235.62) | 0.431 | 0.78 | -0.35 | DOWN |
| Butyric acid | 7.68 (4.44) | 9.1 (6.04) | 0.494 | 1.19 | 0.25 | UP |
| Carnosine | 11 (3.75) | 10.82 (5.46) | 0.561 | 0.98 | -0.02 | DOWN |
| Choline | 57.42 (39.46) | 74.23 (109.13) | 0.633 | 1.29 | 0.37 | UP |
| Citric acid | 321.2 (230.27) | 609.29 (1345.26) | 0.678 | 1.90 | 0.92 | UP |
| Creatine | 5,900.8 (3,108.32) | 5826.27 (3548.61) | 0.983 | 0.99 | -0.02 | DOWN |
| Creatinine | 1,440.00 (0.00) | 1,440.00 (0.00) | 1.000 | 1.00 | 0.00 | --- |
| Hippuric acid | 17,384.67 (5,496.79) | 19,588.00 (5,948.72) | 0.245 | 1.13 | 0.17 | UP |
| Histamine | 0.07 (0.04) | 0.07 (0.06) | 0.868 | 1.10 | 0.13 | UP |
| Homovanillic acid | 8.93 (4.04) | 9.72 (5.95) | 0.934 | 1.09 | 0.12 | UP |
| Indole acetic acid | 33.97 (17.46) | 36.08 (12.41) | 0.361 | 1.06 | 0.09 | UP |
| Isobutyric acid | 3.03 (1.40) | 7.91 (8.13) | 0.320 | 2.61 | 1.38 | UP |
| Kynurenine | 0.74 (0.18) | 0.76 (0.41) | 0.772 | 1.03 | 0.04 | UP |
| Lactic acid | 85.64 (34.87) | 119.79 (141.47) | 0.803 | 1.40 | 0.48 | UP |
| Methionine-sulfoxide | 3.15 (1.18) | 3.15 (1.91) | 0.619 | 1.00 | 0.00 | --- |
| Methylhistidine | 249.20 (60.61) | 292.11 (137.63) | 0.263 | 1.17 | 0.23 | UP |
| Methylmalonic acid | 10.78 (5.42) | 16.54 (9.67) | 0.062 | 1.53 | 0.62 | UP |
| p-Hydroxyhippuric acid | 46.69 (33.28) | 47.43 (53.63) | 0.619 | 1.02 | 0.02 | UP |
| Putrescine | 1.14 (1.62) | 0.82 (0.92) | 0.455 | 0.71 | -0.49 | DOWN |
| Pyruvic acid | 6.27 (2.64) | 11.81 (12.14) | 0.020 | 1.89 | 0.91 | UP |
| Sarcosine | 7.74 (5.35) | 5.90 (4.68) | 0.281 | 0.76 | -0.39 | DOWN |
| Serotonin | 1.40 (0.33) | 1.53 (0.75) | 1.000 | 1.09 | 0.13 | UP |
| Spermidine | 0.11 (0.07) | 0.13 (0.07) | 0.431 | 1.17 | 0.23 | UP |
| Spermine | 0.12 (0.05) | 0.11 (0.05) | 0.917 | 0.97 | -0.04 | DOWN |
| Succinic acid | 24.74 (21.09) | 23.28 (25.5) | 0.481 | 0.94 | -0.09 | DOWN |
| Taurine | 425.51 (359.00) | 370.12 (180.44) | 0.803 | 0.87 | -0.20 | DOWN |
| TDMA | 27.73 (7.04) | 31.63 (15.98) | 0.803 | 1.14 | 0.19 | UP |
| trans-Hydroxyproline | 2.03 (0.90) | 2.13 (1.67) | 0.648 | 1.05 | 0.07 | UP |
| Trimethylamine N-oxide | 1,837.73 (1,663.15) | 2,385.80 (1,881.73) | 0.407 | 1.30 | 0.38 | UP |
| Tyramine | 0.10 (0.06) | 0.11 (0.07) | 0.633 | 1.11 | 0.15 | UP |
| Uric acid | 3,133.33 (1,041.73) | 4,234.00 (2,610.31) | 0.351 | 1.35 | 0.43 | UP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





