Submitted:
01 December 2024
Posted:
02 December 2024
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Collection of Soil Sample
2.2. Preparation of Medium for Isolation of Fungal Species
2.3. Preparation of Potato Dextrose Agar for Isolation of Fungal Species
2.4. Isolation and Sub-Culturing of Fungal Species
2.5. Identification of Fungal Isolates
2.6. Qualitative (Primary) Screening for Pectinolytic Fungal Species
2.7. Quantitative (Secondary) Screening for Pectinolytic Fungal Species
3. Results and Discussion
3.1. Isolation and Sub-Culturing of Fungal Species
3.2. Identification of Fungal Isolates
3.3. Qualitative (primary) screening for pectinolytic fungal species
3.4. Quantitative screening of potent pectinolytic fungal species (secondary screening)
4. Conclusions
Author Contributions
Funding
References
- Shilpa, M.K.; Jason, Y. Isolation and screening of pectinase-producing bacteria from soil sample. CGC Int. j. contemp. res. eng. technol. 2021, 3, 166–170. [Google Scholar]
- Belkheiri, A.; Forouhar, A.; Ursu, A.V.; Dubessay, P.; Pierre, G.; Delattre, C.; Djelveh, G.; Abdelkafi, S.; Hamdami, N.; Michaud, P. Extraction, Characterization, and Applications of Pectins from Plant By-Products. Appl. Sci. 2021, 11, 6596. [Google Scholar] [CrossRef]
- Rokade, D.D.; Vaidya, S.L.; Naziya, M.A.; Dixit, P.P. Screening of Pectinase Producing Bacteria, Isolated from Osmanabad fruit market soil. Int. J. Interdiscip. Multidiscip. 2015, 2, 141–141. [Google Scholar]
- Tabssum, F.; Ali, S.S. Screening of Pectinase Producing Gram Positive Bacteria: Isolation and Characterization. Punjab- Univ. J. Zoöl. 2018, 33. [Google Scholar] [CrossRef]
- Thakur, P.; Singh, A.K.; Mukherjee, G. Isolation and Characterization of Alkaline Pectinase Productive Bacillus tropicus from Fruit and Vegetable Waste Dump Soil. Braz. Arch. Biol. Technol. 2021, 64. [Google Scholar] [CrossRef]
- Rakitikul, W.; Tammasat, T.; Udjai, J.; Nimmanpipug, P. Experimental and DFT study of gelling factor of pectin. Suranaree J. Sci. Technol. 2016, 23, 421–428. [Google Scholar]
- Food Science Avenue (2016): Pectinic acid. Pectinic-acid.html (Date assessed: 20/02/2023).
- Freitas, C.M.P.; Coimbra, J.S.R.; Souza, V.G.L.; Sousa, R.C.S. Structure and Applications of Pectin in Food, Biomedical, and Pharmaceutical Industry: A Review. Coatings 2021, 11, 922. [Google Scholar] [CrossRef]
- Lucarini, M.; Durazzo, A.; Bernini, R.; Campo, M.; Vita, C.; Souto, E.B.; Lombardi-Boccia, G, Ramadan; Santini, A.; Romani, A. Fruit Wastes as a Valuable Source of Value-Added Compounds: A Collaborative Perspective. Molecules 2021, 26, 6338. [Google Scholar] [CrossRef]
- Chandel, Vinay, Deblina Biswas, Swarup Roy, Devina Vaidya, Anil Verma, and Anil Gupta. (2022). “Current Advancements in Pectin: Extraction, Properties and Multifunctional Applications” Foods 11, no. 17: 2683.
- Tan, J.; Hua, X.; Liu, J.; Wang, M.; Liu, Y.; Yang, R.; Cao, Y. Extraction of sunflower head pectin with superfine grinding pretreatment. Food Chem. 2020, 320, 126631. [Google Scholar] [CrossRef]
- Picot-Allain, M.C.N.; Ramasawmy, B.; Emmambux, M.N. Extraction, Characterisation, and Application of Pectin from Tropical and Sub-Tropical Fruits: A Review. Food Rev. Int. 2020, 38, 282–312. [Google Scholar] [CrossRef]
- Roman-Benn, A.; Contador, C.A.; Li, M.-W.; Lam, H.-M.; Ah-Hen, K.; Ulloa, P.E.; Ravanal, M.C. Pectin: An overview of sources, extraction and applications in food products, biomedical, pharmaceutical and environmental issues. Food Chem. Adv. 2023, 2. [Google Scholar] [CrossRef]
- Hassan, S. (2020). Development of novel Pectinase and Xylanase juice clarifcation enzymes via a combined biorefnery and immobilization approach, Doctoral Thesis, Technological University Dublin. [CrossRef]
- Baljinder Singh Kauldhar, Harpreet Kaur, Venkatesh Meda, Balwinder Singh Sooch (2022). Chapter 12 - Insights into upstreaming and downstreaming processes of microbial extremozymes. Extremozymes and Their Industrial Applications, Academic Press, Pages 321-352, ISBN 9780323902748. [CrossRef]
- Shet AR, Desai SV, Achappa S. (2018). Pectinolytic enzymes: classification, production, purification and applications. Res. J. Life Sci Bioinform Pharm Chem Sci.; 4:337 –48.
- Soorej M. Basheer, Sreeja Chellappan, A. Sabu (2022). Chapter 8 - Enzymes in fruit and vegetable processing, Editor(s): Mohammed Kuddus, Cristobal Noe Aguilar, Value-Addition in Food Products and Processing through Enzyme Technology, Academic Press, Pages 101-110, ISBN 9780323899291.
- Garg, G.; Singh, A.; Kaur, A.; Singh, R.; Kaur, J.; Mahajan, R. Microbial pectinases: an ecofriendly tool of nature for industries. 3 Biotech 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Oumer, O.J.; Abate, D. Screening and Molecular Identification of Pectinase Producing Microbes from Coffee Pulp. BioMed Res. Int. 2018, 2018, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Satapathy, S.; Behera, P.M.; Tanty, D.K.; Srivastava, S.; Thatoi, H.; Dixit, A.; Sahoo, S.L. Isolation and molecular identification of pectinase producing Aspergillus species from different soil samples of Bhubaneswar regions. BioRxiv 2019, 1, 1–22. [Google Scholar]
- Sarita Shrestha; Chonlong Chio; Janak Raj Khatiwada; Aristide Laurel Mokale Kognou; Xuantong Chen; Wensheng Qin (2023). Optimization of Cultural Conditions for Pectinase Production by Streptomyces sp. and Characterization of Partially Purified Enzymes. Microb Physiol 33 (1): 12–26. PubMed: 36417846.
- Xiangyang Liu, Chandrakant Kokare, (2023). Chapter 17 - Microbial enzymes of use in industry, Editor(s): Goutam Brahmachari, Biotechnology of Microbial Enzymes (Second Edition), Academic Press, Pages 405-444, ISBN 9780443190599.
- Osete-Alcaraz, A.; Gómez-Plaza, E.; Pérez-Porras, P.; Bautista-Ortín, A.B. Revisiting the use of pectinases in enology: A role beyond facilitating phenolic grape extraction. Food Chem. 2022, 372, 131282. [Google Scholar] [CrossRef]
- Haile, S.; Ayele, A. Pectinase from Microorganisms and Its Industrial Applications. Sci. World J. 2022, 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nazneen Inamdar, Asmita Dike, Amol Jadhav (2024). Isolation, Screening, and Identification of Pectin Degrading Bacteria from Soil. Journal of Chemical Health Risks, JCHR 14(2), 930-938 | ISSN: 2251-6727.
- Ivarsson, M.; Drake, H.; Bengtson, S.; Rasmussen, B. A Cryptic Alternative for the Evolution of Hyphae. BioEssays 2020, 42, e1900183. [Google Scholar] [CrossRef]
- Oliyad, J.O., Dawit, A. (2018). Screening and molecular identification of pectinase producing microbes from coffee pulp. Biomed Research International: 2961767.
- Carlos, D.L.-M.J.; Leonardo, S.; Jesús, M.-C.; Paola, M.-R.; Alejandro, Z.-C.; Juan, A.-V.; Noé, A.C. Solid-State Fermentation with Aspergillus niger GH1 to Enhance Polyphenolic Content and Antioxidative Activity of Castilla Rose (Purshia plicata). Plants 2020, 9, 1518. [Google Scholar] [CrossRef]
- Sudeep, K.C, Jitendra U. Dev R. J., Binod L, Dhiraj K. C., Bhoj R. P, Tirtha R. B., Rajiv D. Santosh K., Niranjan K., 10,Vijaya R. (2020). Production, Characterization, and Industrial Application of Pectinase Enzyme Isolated from Fungal Strains. Fermentation, 6, 59; 6.
- Singh, A.; Varghese, L.M.; Yadav, R.D.; Mahajan, R. A pollution reducing enzymatic deinking approach for recycling of mixed office waste paper. Environ. Sci. Pollut. Res. 2020, 27, 45814–45823. [Google Scholar] [CrossRef] [PubMed]
- Okonji, R.E.; Itakorode, B.O.; Ovumedia, J.O.; Adedeji, O.S. Purification and biochemical characterization of pectinase produced by Aspergillus fumigatus isolated from soil of decomposing plant materials. Journal of Applied Biology & Biotechnology 2019, 7, 1–8. [Google Scholar]
- Mrinmoy Ghosh and Krishna Kanth Pulicherla (2021). Psychrophiles as the source for potential industrial psychrozymes. Recent Developments in Microbial Technologies, Environmental and Microbial Biotechnology. [CrossRef]
- Ram Prasad, Vivek Kumar, Joginder Singh, Chandrama Prakash Upadhyaya (2021). Recent Developments in Microbial Technologies. Environmental and Microbial Biotechnology. ISSN 2662-169X.
- Alexopoulos, C.J., Mims, C.W., Blackwell, M. (1996). Intro. Mycology. Vol. 4. New York: Wiley.
- Barnett, H.L.; Hunter, B.B. Illustrated Genera of Imperfect Fungi, 4th ed.; Practice Hall: New York, NY, USA, 1987. [Google Scholar]
- Kaur, H.P.; Kaur, G. Optimization of cultural conditions for pectinase produced by fruit spoilage fungi. International journal of advances in research 2014, 3, 851–859. [Google Scholar]
- Adedayo, M.R.; Mohammed, M.T.; Ajiboye, A.E.; A Abdulmumini, S. Pectinolytic activity of aspergillus niger and Aspergillus flavus grown on grapefruit (citrus Parasidis) peel in solid state fermentation. Glob. J. Pure Appl. Sci. 2021, 27, 93–105. [Google Scholar] [CrossRef]
- El-Ghomary, A.E.; Shoukry, A.A.; El-Kotkat, M.B. Productivity of pectinase enzymes by Aspergillus sp. isolated from Egyptian soil. Al-Azhar J. Agric. Res. 2021, 46, 79–87. [Google Scholar] [CrossRef]
- Martison Budu, Patrick Boakye, Joseph Asankomah Bentil (2023). Microbial diversity, enzyme profile and substrate concentration for bioconversion of cassava peels to bioethanol. Preprints. [CrossRef]
- Timothy C. Cairns, Corrado Nai and Vera Meyer (2018). Aspergillus niger: A Hundred Years of Contribution to the Natural Products Chemistry. Fungal Biol Biotechnol, 5:13.
- Ezenwelu, C.O.; Afeez, O.A.; Anthony, O.U.; Promise, O.A.; Mmesoma, U.-E.C.; Henry, O.E. Studies on Properties of Lipase Produced from Aspergillus sp. Isolated from Compost Soil. Adv. Enzym. Res. 2022, 10, 49–60. [Google Scholar] [CrossRef]
- Ametefe, G.D., Oluwadamilare, L.A., Ifeoma, C. J., Olubunmi, I. I., Ofoegbu, V. O., Folake, F., Orji, F. A., Iweala, E.E.J., and Chinedu, S. N. (2021). Optimization of pectinase activity from locally isolated fungi and agrowastes. Research square.
- Jalil, M.T.M.; Ibrahim, D. Partial Purification and Characterisation of Pectinase Produced by Aspergillus niger LFP-1 Grown on Pomelo Peels as a Substrate. Trop. Life Sci. Res. 2021, 32, 1–22. [Google Scholar] [CrossRef]
- Oumer, O.J. Pectinase: Substrate, Production and their Biotechnological Applications. Int. J. Environ. Agric. Biotechnol. 2017, 2, 1007–1014. [Google Scholar] [CrossRef]
- Kamalambigeswari, R.; Sangilimuthu, A.Y.; Narender, S.; Ushani, U. Isolation, identification, screening and optimization of pectinase producing soil fungi (Aspergillus niger). International Journal of Research in Pharmaceutical Sciences 2018, 9, 762–762. [Google Scholar]

| Isolate | Characteristics (Macro-and Micromorphological) | Possible Identity |
|---|---|---|
|
FS1 |
Appeared white initially and later turned grey or yellowish brown whilst pale yellow appeared as the reverse; colonies were fast growing with the texture appearing velvety or powdery, with a cottony appearance; conidial heads appeared as compact clusters, typically yellowish-green to yellow in color; large spore-bearing heads, spherical, and densely packed; clear, thin, and short hyphae; greenish mycelia; branched greenish conidia and septate hyphae; dense unbranched conidiophores with rounded or flask-shaped vesicle; elongated Phialides with constricted neck. |
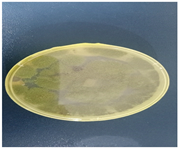 Aspergillus flavus Aspergillus flavus
|
|
FS2 |
Colony had fast and aggressive growth and looked fluffy white initially, turned black later; generated enormous black spores; reverse appeared pale yellow; wrinkled mycelia with visible dense and lengthy aerial hyphae; conidia were unbranched; texture looked velvety or powdery and the conidial produced heads with compact clusters, typically dark green to black in colour; hyphae looked septate, dense and branched; long, unbranched, and terminate conidiophores and gave rise to the conidial heads; rounded or flask-shaped vesicle at the end of conidiophore; Phialides were elongated with constricted neck. |
 Aspergillus niger. |
|
FS3 |
Colony was fast-growing and dominant with white appearance; sticky green phialoconidia clusters formed within a few days; flask-like Phialides with an enlarged base; woolly colonies formed initially and got compacted over time; as the conidia developed, dispersed blue green or yellow-green spots appeared visible, the reverse was pale, tan, or yellowish; conidia usually appeared within a week in dense or loose tufts of green, yellow, or white.; a yellow pigment was observed to have been released into the agar medium; an irregular yellow zone without conidia was present around the colony; Some concentric circles/rings also appeared. |
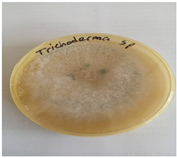 Trichoderma spp. Trichoderma spp. |
|
FS4 |
Colonies grew very fast, and looked pale or brightly-coloured with the mycelia appearing like cotton; the thallus looked whitish in colour and showed presence of conidia from slender phialides; conidiophores looked solitary, short, lateral monophialides in the airborne mycelium, but were later observed as thickly branching groups; macroconidia looked fusiform, slightly curved and pointed at the tip and also appeared plentiful, largely non-septate, and straight or curved and looked cylindrically-shaped; chlamydospores appeared terminal, smooth and rough-walled; the phialides were short and mainly non-septate. |
 Fusarium spp. Fusarium spp. |
|
FS5 |
Colonies of Saccharomyces grew rapidly and matured in three days; they looked small, flat, smooth, moist and creamy or white in colour, raised and clustered, exhibiting oval or spherical to ellipsoidal shapes. |
 Saccharomyces spp. Saccharomyces spp. |
| Microorganism | Zone of Pectinolytic activity/mm |
|---|---|
| Aspergillus niger | 25.00 |
| Fusarium spp. | 23.00 |
| Trichoderma spp. | 20.00 |
| Aspergillus flavus | 20.00 |
| Saccharomyces spp. | 7.00 |
| P-value = 0.0511. | |
| Microorganism | Reducing sugar (mg/mL) | Enzyme activity (U/mL) |
|---|---|---|
| Aspergillus niger (FS2) | 3.92 | 36.23 |
| Aspergillus flavus (FS1) | 3.68 | 32.52 |
| Trichoderma spp. (FS3) | 3.23 | 29.74 |
| Fusarium spp. (FS4) | 3.16 | 28.66 |
| P-value = 0.0260. | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




