Submitted:
10 April 2025
Posted:
12 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Impact of CCFA on the Lamb’s Body Weight
2.2. Whole Genome Shotgun Sequencing Summary
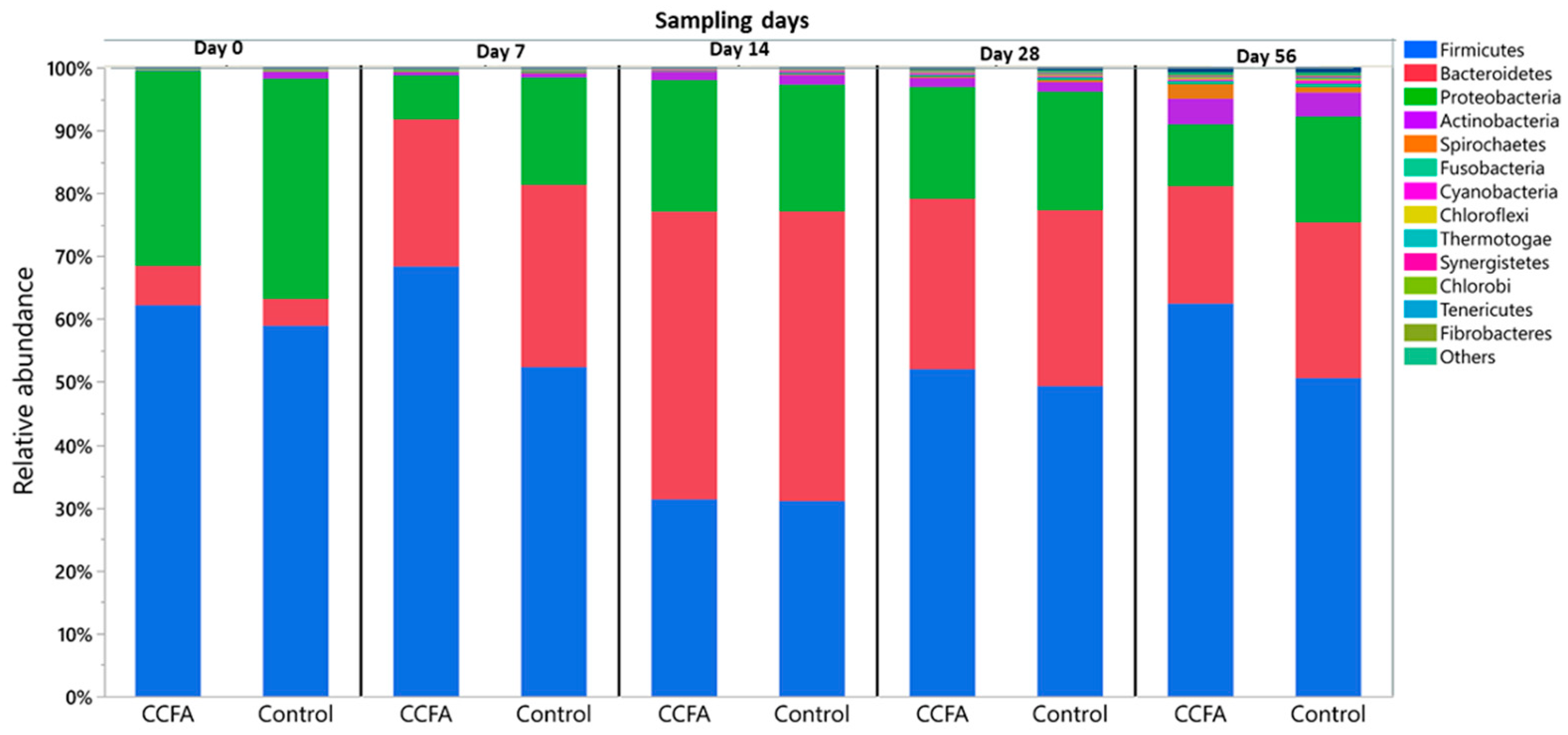
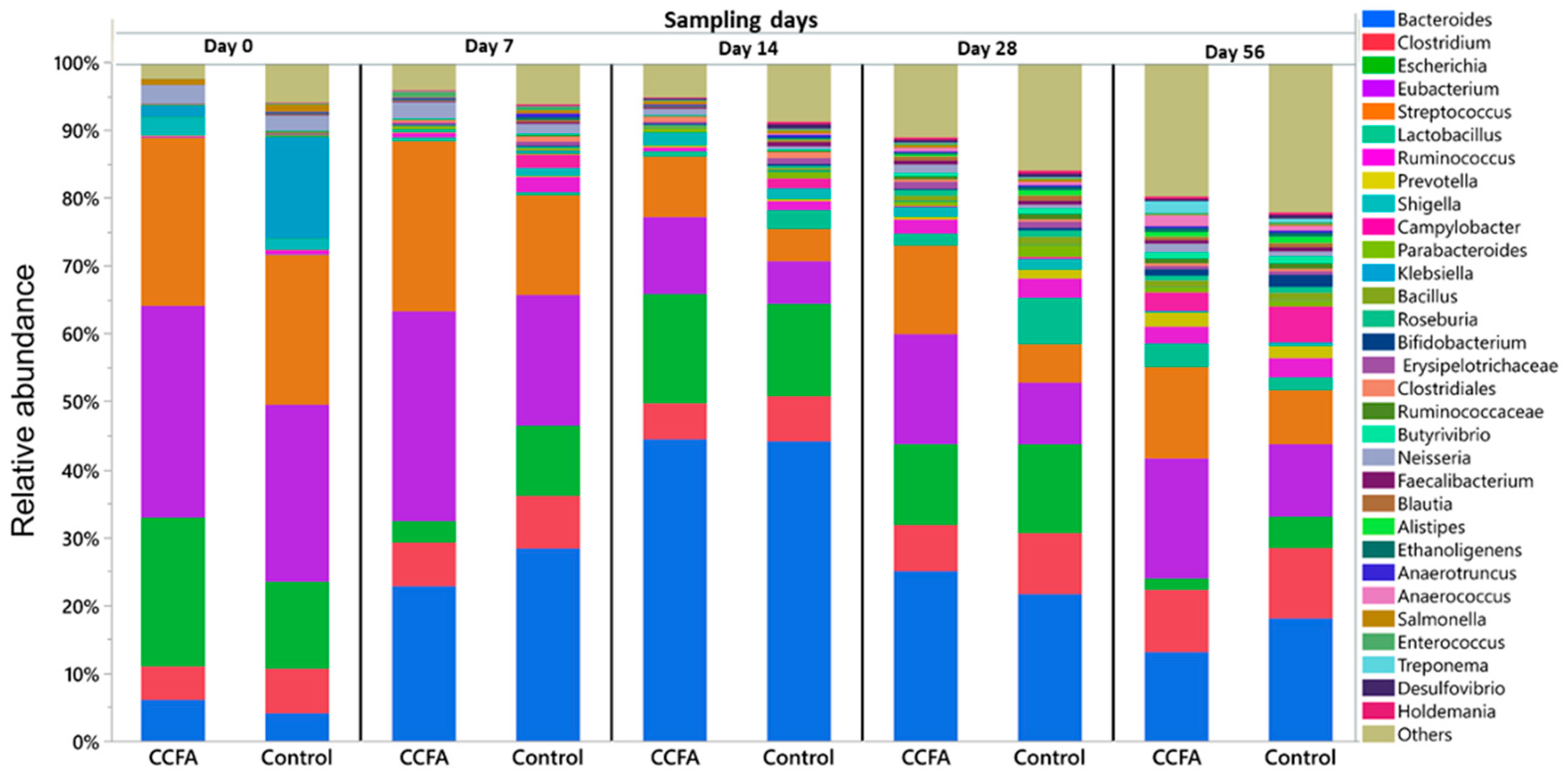
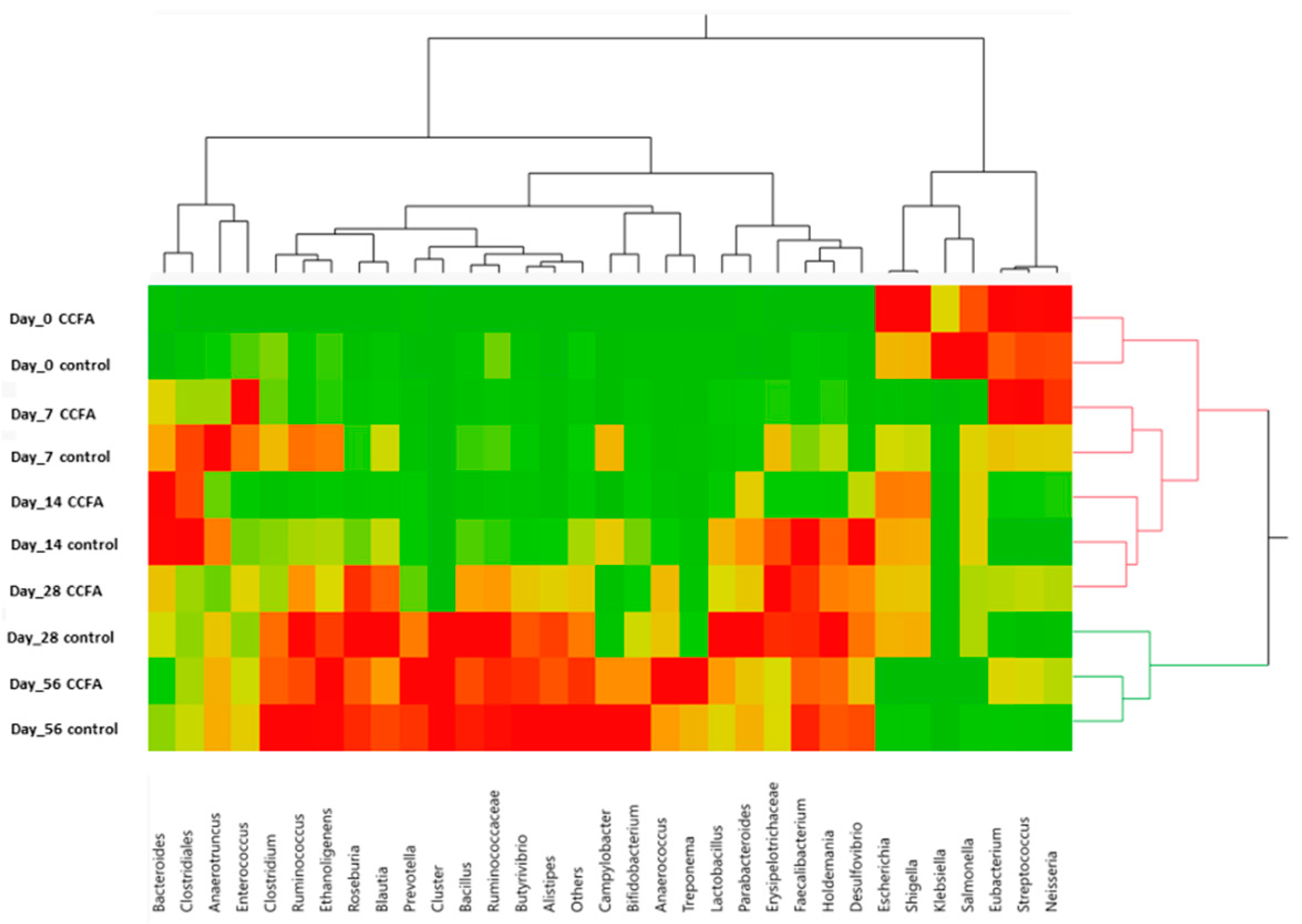
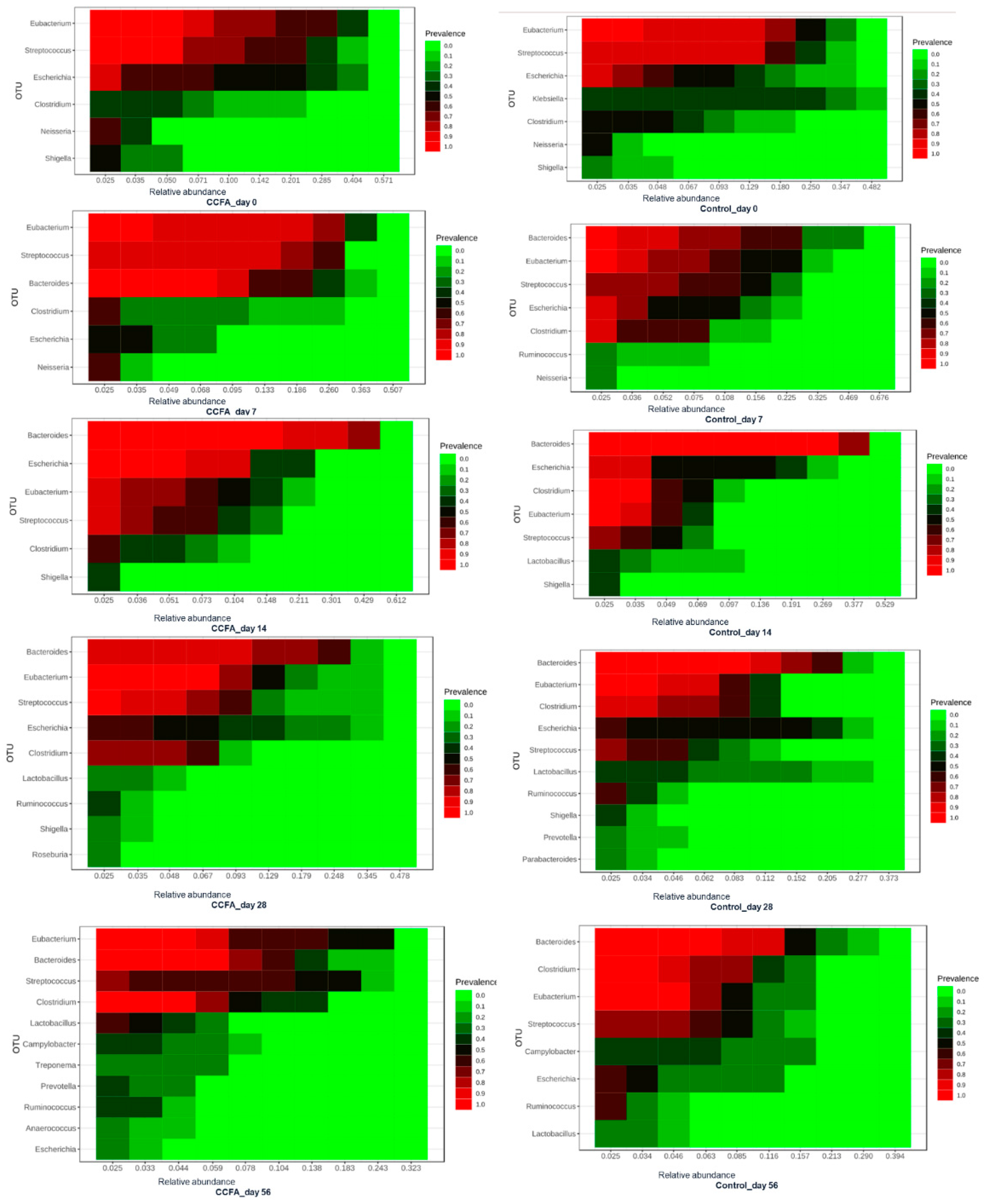
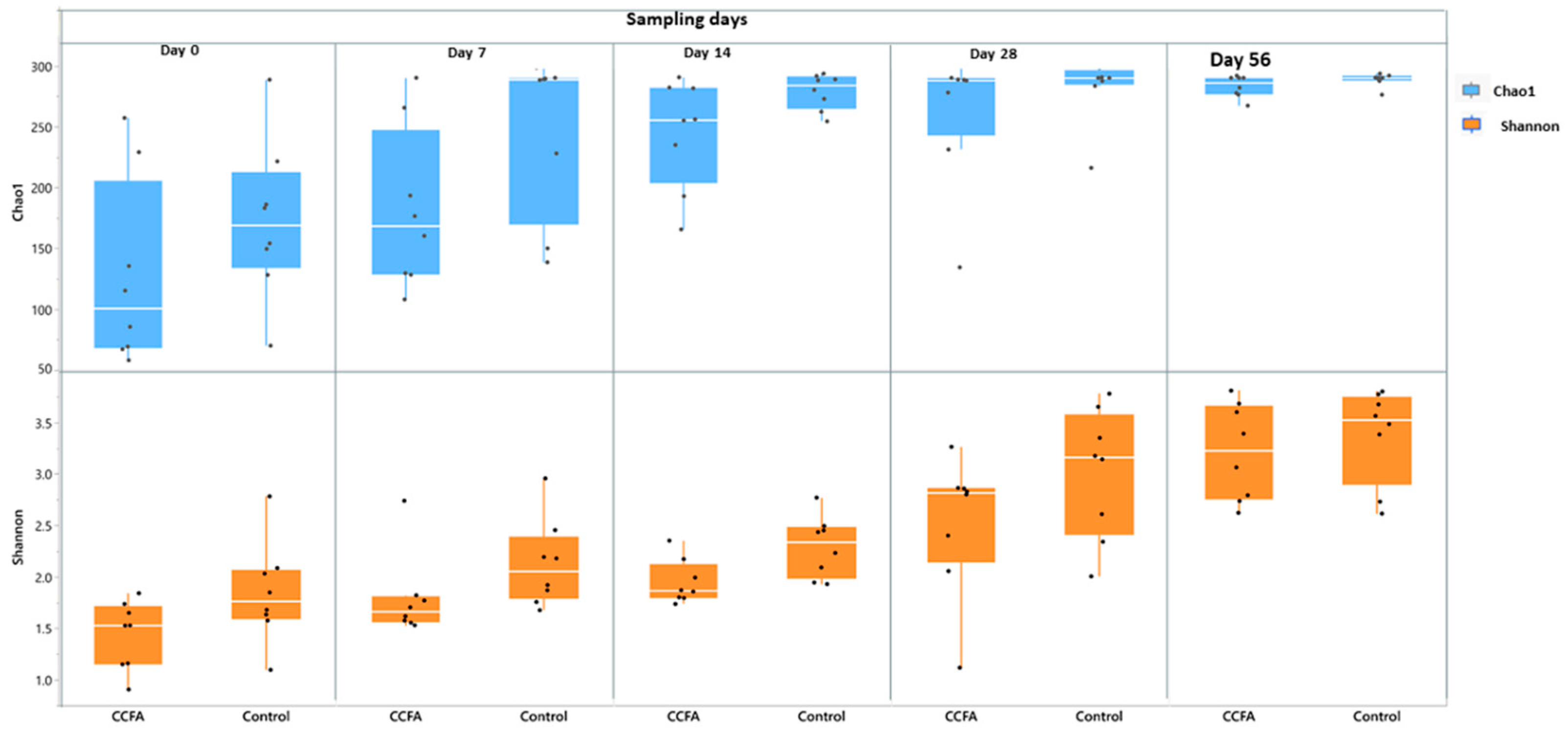
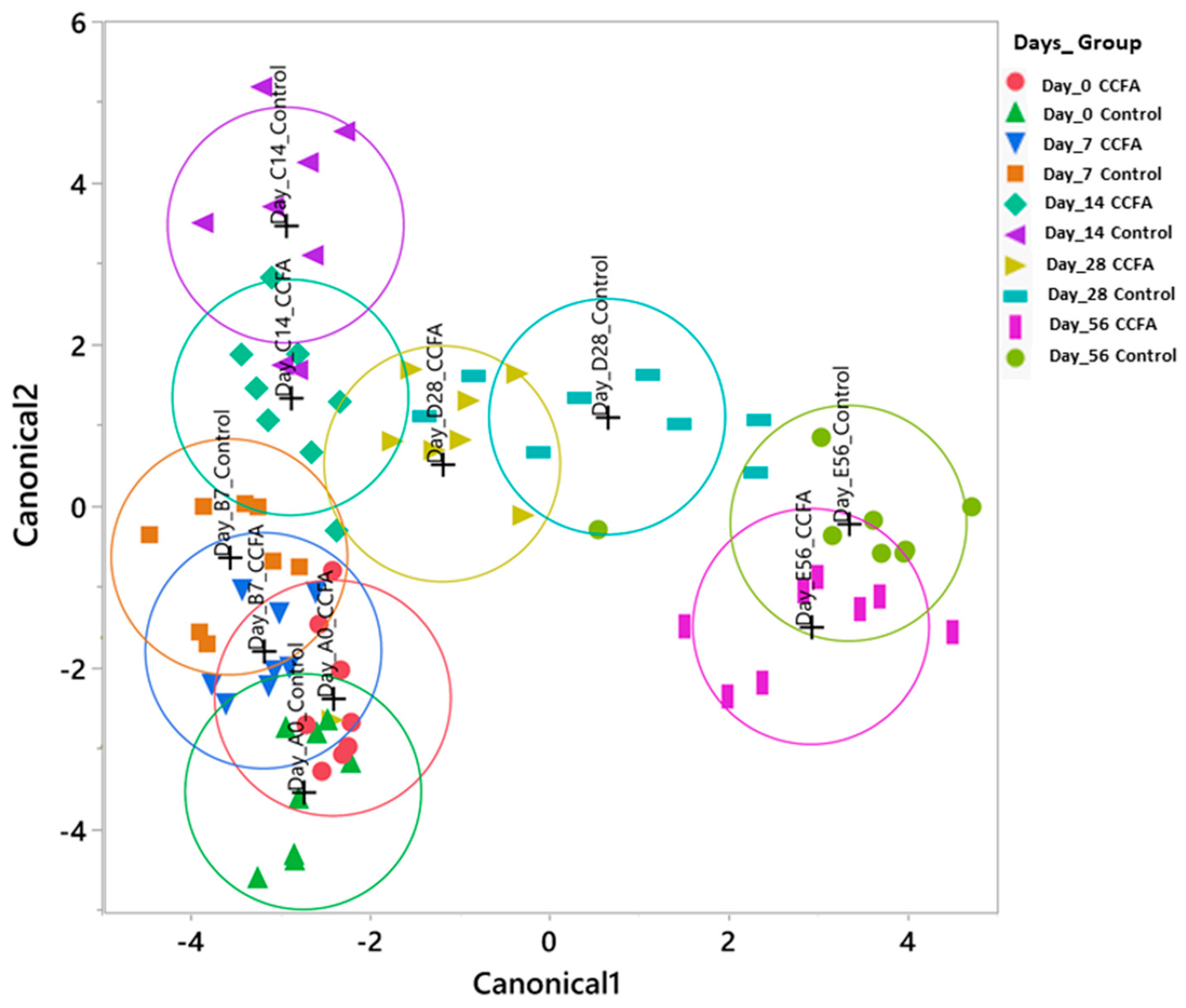
2.3. Effects of the Early Ceftiofur Treatment on the Lamb Fecal Microbiome Composition and Diversity
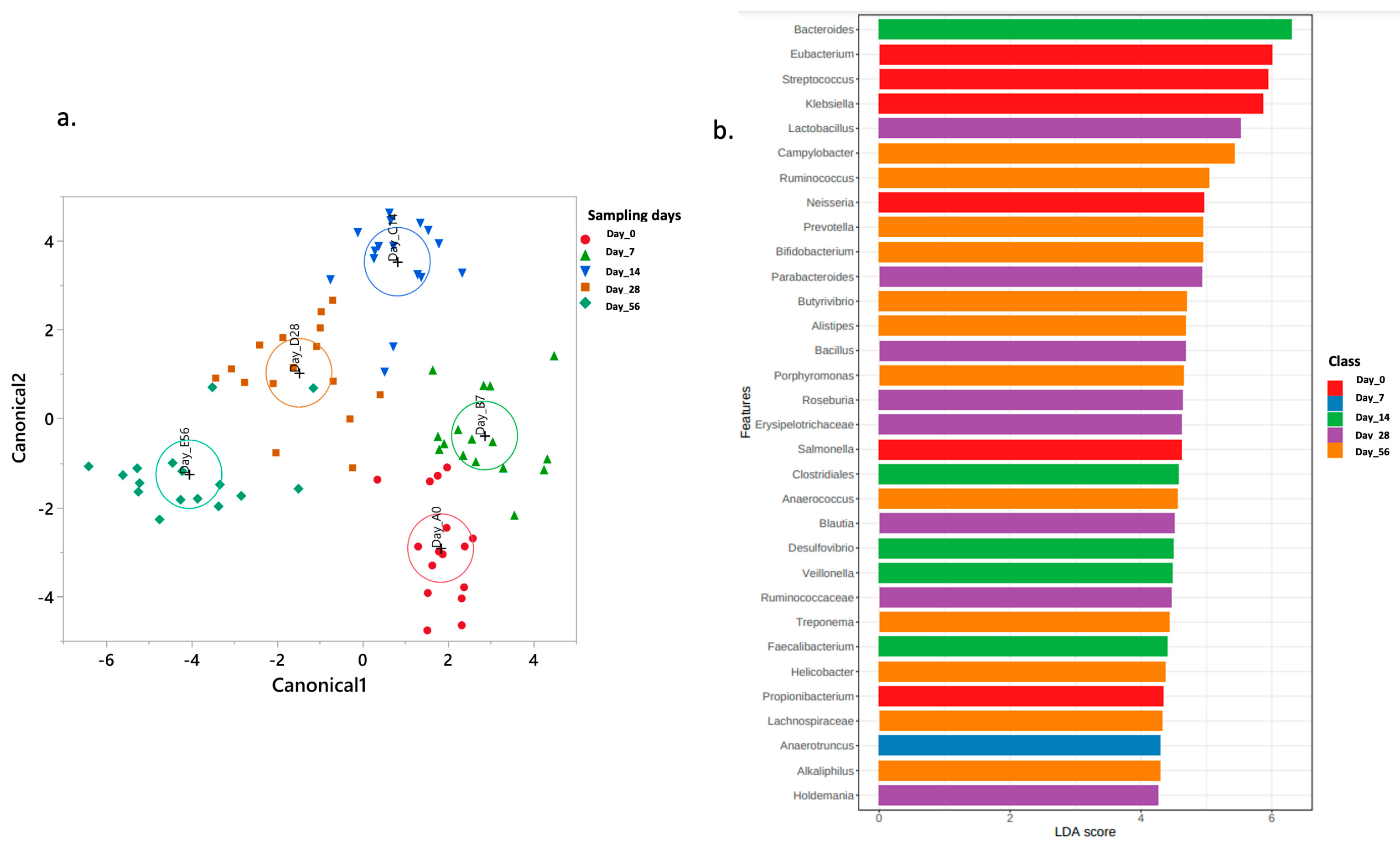
2.4. Normal Fecal Microbiota Development
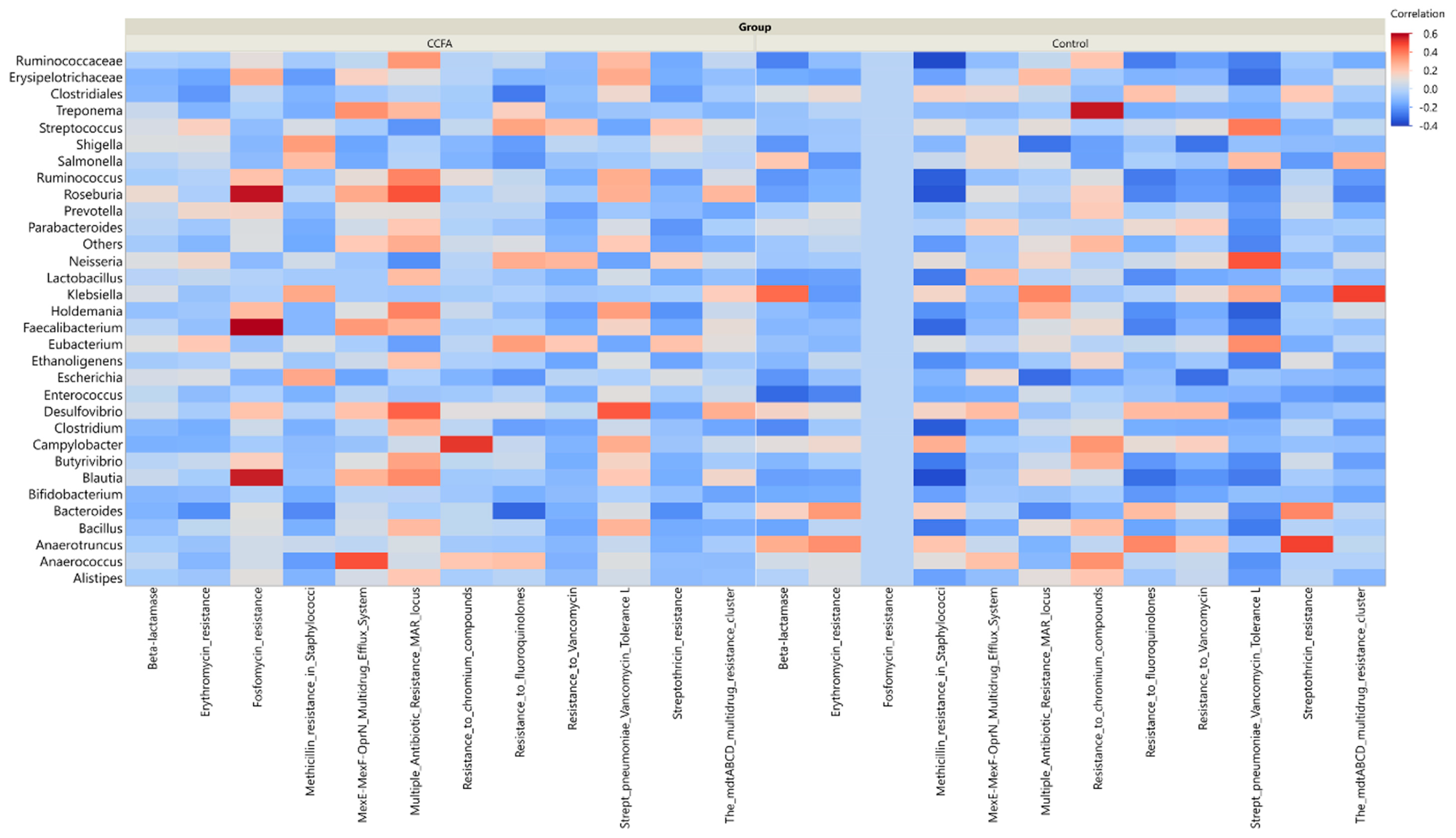
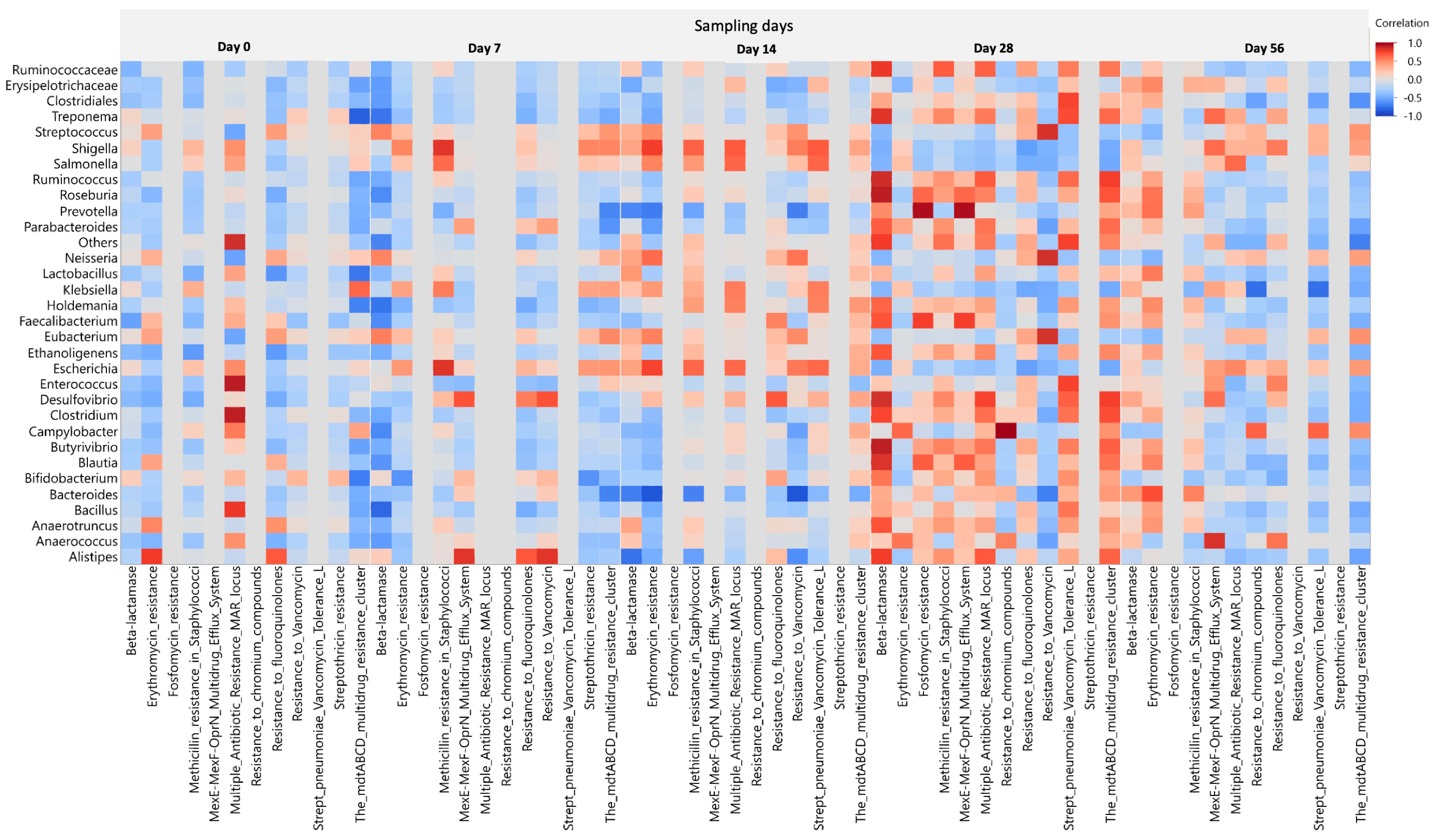
2.5. Effect of Ceftiofur on the Microbial Functional Resistant Genes
3. Discussion
4. Materials and Methods
4.1. Experimental Design and Sample Collection
4.1.1. Animals
4.1.2. Challenge
4.1.3. Samples
4.1.4. Ethics Statement of Animal Use
4.2. DNA Extraction
4.3. Whole Genome Shotgun DNA Sequencing (WGSS)
4.4. Bioinformatic Analysis
4.4.1. Sequence Reads Processing
4.4.2. Operational Taxonomic Units (OTUs) Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CCFA | Ceftiofur Crystalline Free Acid |
| ARG(s) | Antibiotic Resistance Gene(s) |
| WGSS | Whole Genome Shotgun Sequencing |
| DNA | Deoxyribonucleic Acid |
| OTUs | Operational Taxonomic Units |
| MG-RAST | Metagenomics Rapid Annotation using Subsystem Technology |
| RefSeq | Reference Sequence Database |
| PCA | Principal Component Analysis |
| CDA | Canonical Discriminant Analysis |
| LDA | Linear Discriminant Analysis |
| LEfSe | Linear Discriminant Analysis Effect Size |
| PCR | Polymerase Chain Reaction |
References
- Alonso, V.R.; Guarner, F. Linking the gut microbiota to human health. British Journal of Nutrition 2013, 109, S21–S26. [Google Scholar] [CrossRef] [PubMed]
- Malmuthuge, N.; Griebel, P.J.; Guan, L.L. The gut microbiome and its potential role in the development and function of newborn calf gastrointestinal tract. Frontiers in veterinary science 2015, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- van den Broek, M.F.; De Boeck, I.; Kiekens, F.; Boudewyns, A.; Vanderveken, O.M.; Lebeer, S. Translating recent microbiome insights in otitis media into probiotic strategies. Clinical microbiology reviews 2019, 32, e00010–00018. [Google Scholar] [CrossRef]
- Khalil, A.; Batool, A.; Arif, S. Healthy Cattle Microbiome and Dysbiosis in Diseased Phenotypes. Ruminants 2022, 2, 134–156. [Google Scholar] [CrossRef]
- Kraimi, N.; Dawkins, M.; Gebhardt-Henrich, S.G.; Velge, P.; Rychlik, I.; Volf, J.; Creach, P.; Smith, A.; Colles, F.; Leterrier, C. Influence of the microbiota-gut-brain axis on behavior and welfare in farm animals: A review. Physiology & behavior 2019, 210, 112658. [Google Scholar]
- Guo, C.Y.; Ji, S.K.; Yan, H.; Wang, Y.J.; Liu, J.J.; Cao, Z.J.; Yang, H.J.; Zhang, W.J.; Li, S.L. Dynamic change of the gastrointestinal bacterial ecology in cows from birth to adulthood. MicrobiologyOpen 2020, 9, e1119. [Google Scholar] [CrossRef] [PubMed]
- Li, R.W.; Connor, E.E.; Li, C.; Baldwin, R.L., VI; Sparks, M.E. Characterization of the rumen microbiota of pre-ruminant calves using metagenomic tools. Environmental microbiology 2012, 14, 129–139. [Google Scholar] [CrossRef]
- Oikonomou, G.; Teixeira, A.G.V.; Foditsch, C.; Bicalho, M.L.; Machado, V.S.; Bicalho, R.C. Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. Associations of Faecalibacterium species with health and growth. PloS one 2013, 8, e63157. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Griebel, P.J.; Guan, L.L. Taxonomic identification of commensal bacteria associated with the mucosa and digesta throughout the gastrointestinal tracts of preweaned calves. Applied and environmental microbiology 2014, 80, 2021–2028. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Li, M.; Chen, Y.; Fries, P.; Griebel, P.J.; Baurhoo, B.; Zhao, X.; Guan, L.L. Distinct commensal bacteria associated with ingesta and mucosal epithelium in the gastrointestinal tracts of calves and chickens. FEMS Microbiology Ecology 2012, 79, 337–347. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The gut microbiota—masters of host development and physiology. Nature reviews microbiology 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Janssen, P.H. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Scientific reports 2015, 5, 1–15. [Google Scholar] [CrossRef]
- Weimer, P.; Stevenson, D.; Mantovani, H.; Man, S. Host specificity of the ruminal bacterial community in the dairy cow following near-total exchange of ruminal contents. Journal of dairy science 2010, 93, 5902–5912. [Google Scholar] [CrossRef]
- Benson, A.K.; Kelly, S.A.; Legge, R.; Ma, F.; Low, S.J.; Kim, J.; Zhang, M.; Oh, P.L.; Nehrenberg, D.; Hua, K. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proceedings of the National Academy of Sciences 2010, 107, 18933–18938. [Google Scholar] [CrossRef]
- Blaser, M.J. Antibiotic use and its consequences for the normal microbiome. Science 2016, 352, 544–545. [Google Scholar] [CrossRef]
- Sawant, A.; Sordillo, L.; Jayarao, B. A survey on antibiotic usage in dairy herds in Pennsylvania. Journal of dairy science 2005, 88, 2991–2999. [Google Scholar] [CrossRef]
- Wang, C.; Wei, S.; Chen, N.; Xiang, Y.; Wang, Y.; Jin, M. Characteristics of gut microbiota in pigs with different breeds, growth periods and genders. Microbial biotechnology 2022, 15, 793–804. [Google Scholar] [CrossRef]
- Kobayashi, R.; Nagaoka, K.; Nishimura, N.; Koike, S.; Takahashi, E.; Niimi, K.; Murase, H.; Kinjo, T.; Tsukahara, T.; Inoue, R. Comparison of the fecal microbiota of two monogastric herbivorous and five omnivorous mammals. Animal science journal 2020, 91, e13366. [Google Scholar] [CrossRef]
- Esteban-Blanco, C.; Gutiérrez-Gil, B.; Marina, H.; Pelayo, R.; Suárez-Vega, A.; Acedo, A.; Arranz, J.-J. The milk microbiota of the spanish churra sheep breed: New insights into the complexity of the milk microbiome of dairy species. Animals 2020, 10, 1463. [Google Scholar] [CrossRef]
- Pantoja-Feliciano, I.G.; Clemente, J.C.; Costello, E.K.; Perez, M.E.; Blaser, M.J.; Knight, R.; Dominguez-Bello, M.G. Biphasic assembly of the murine intestinal microbiota during early development. The ISME journal 2013, 7, 1112–1115. [Google Scholar] [CrossRef]
- MINATO, H.; OTSUKA, M.; SHIRASAKA, S.; ITABASHI, H.; MITSUMORI, M. Colonization of microorganisms in the rumen of young calves. The Journal of General and Applied Microbiology 1992, 38, 447–456. [Google Scholar] [CrossRef]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. The ISME journal 2013, 7, 1069–1079. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef]
- Arshad, M.A.; Hassan, F.-u.; Rehman, M.S.; Huws, S.A.; Cheng, Y.; Din, A.U. Gut microbiome colonization and development in neonatal ruminants: Strategies, prospects, and opportunities. Animal Nutrition 2021, 7, 883–895. [Google Scholar] [CrossRef]
- O’Mahony, L.; McCarthy, J.; Kelly, P.; Hurley, G.; Luo, F.; Chen, K.; O’Sullivan, G.C.; Kiely, B.; Collins, J.K.; Shanahan, F. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology 2005, 128, 541–551. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Nagata, R.; Ohtani, N.; Ichijo, T.; Ikuta, K.; Sato, S. Effects of dietary forage and calf starter diet on ruminal pH and bacteria in Holstein calves during weaning transition. Frontiers in microbiology 2016, 7, 1575. [Google Scholar] [CrossRef]
- Kim, E.-T.; Lee, S.-J.; Kim, T.-Y.; Lee, H.-G.; Atikur, R.M.; Gu, B.-H.; Kim, D.-H.; Park, B.-Y.; Son, J.-K.; Kim, M.-H. Dynamic changes in fecal microbial communities of neonatal dairy calves by aging and diarrhea. Animals 2021, 11, 1113. [Google Scholar] [CrossRef]
- Gonzalez-Perez, G.; Hicks, A.L.; Tekieli, T.M.; Radens, C.M.; Williams, B.L.; Lamousé-Smith, E.S. Maternal antibiotic treatment impacts development of the neonatal intestinal microbiome and antiviral immunity. The Journal of Immunology 2016, 196, 3768–3779. [Google Scholar] [CrossRef]
- Hornish, R.E.; Katarski, S. Cephalosporins in veterinary medicine-ceftiofur use in food animals. Current topics in medicinal chemistry 2002, 2, 717–731. [Google Scholar] [CrossRef]
- Song, Y.; Li, F.; Fischer-Tlustos, A.; Neves, A.; He, Z.; Steele, M.; Guan, L. Metagenomic analysis revealed the individualized shift in ileal microbiome of neonatal calves in response to delaying the first colostrum feeding. Journal of Dairy Science 2021, 104, 8783–8797. [Google Scholar] [CrossRef]
- Yeoman, C.J.; Ishaq, S.L.; Bichi, E.; Olivo, S.K.; Lowe, J.; Aldridge, B.M. Biogeographical differences in the influence of maternal microbial sources on the early successional development of the bovine neonatal gastrointestinal tract. Scientific reports 2018, 8, 3197. [Google Scholar] [CrossRef]
- Ruczizka, U.; Metzler-Zebeli, B.; Unterweger, C.; Mann, E.; Schwarz, L.; Knecht, C.; Hennig-Pauka, I. Early parenteral administration of ceftiofur has gender-specific short-and long-term effects on the fecal microbiota and growth in pigs from the suckling to growing phase. Animals 2019, 10, 17. [Google Scholar] [CrossRef]
- Iwiński, H.; Wódz, K.; Chodkowska, K.; Nowak, T.; Różański, H. In vitro evaluation of antimicrobial effect of phytobiotics mixture on Salmonella spp. isolated from chicken broiler. Antibiotics 2022, 11, 868. [Google Scholar] [CrossRef]
- VT Nair, D.; Venkitanarayanan, K.; Kollanoor Johny, A. Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods 2018, 7, 167. [Google Scholar] [CrossRef]
- Deusch, S.; Tilocca, B.; Camarinha-Silva, A.; Seifert, J. News in livestock research—use of Omics-technologies to study the microbiota in the gastrointestinal tract of farm animals. Computational and structural biotechnology journal 2015, 13, 55–63. [Google Scholar] [CrossRef]
- Nowland, T.L.; Torok, V.A.; Low, W.Y.; Barton, M.D.; Plush, K.J.; Kirkwood, R.N. Faecal microbiota analysis of piglets during lactation. Animals 2020, 10, 762. [Google Scholar] [CrossRef]
- Rutjens, S.; Vereecke, N.; De Spiegelaere, W.; Croubels, S.; Devreese, M. Intestinal Exposure to Ceftiofur and Cefquinome after Intramuscular Treatment and the Impact of Ceftiofur on the Pig Fecal Microbiome and Resistome. Antibiotics 2022, 11, 342. [Google Scholar] [CrossRef]
- Candon, S.; Perez-Arroyo, A.; Marquet, C.; Valette, F.; Foray, A.-P.; Pelletier, B.; Milani, C.; Ventura, M.; Bach, J.-F.; Chatenoud, L. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PloS one 2015, 10, e0125448. [Google Scholar] [CrossRef]
- Huda, M.N.; Salvador, A.C.; Barrington, W.T.; Gacasan, C.A.; D’Souza, E.M.; Ramirez, L.D.; Threadgill, D.W.; Bennett, B.J. Gut microbiota and host genetics modulate the effect of diverse diet patterns on metabolic health. Frontiers in Nutrition 2022, 9. [Google Scholar] [CrossRef]
- Thompson, C.L.; Wang, B.; Holmes, A.J. The immediate environment during postnatal development has long-term impact on gut community structure in pigs. The ISME journal 2008, 2, 739–748. [Google Scholar] [CrossRef]
- Schwarzer, M.; Strigini, M.; Leulier, F. Gut microbiota and host juvenile growth. Calcified tissue international 2018, 102, 387–405. [Google Scholar] [CrossRef]
- Liu, L.; Kirst, M.E.; Zhao, L.; Li, E.; Wang, G.P. Microbiome Resilience despite a Profound Loss of Minority Microbiota following Clindamycin Challenge in Humanized Gnotobiotic Mice. Microbiology spectrum 2022, 10, e01960–01921. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS microbiology reviews 2018, 42, fux053. [Google Scholar] [CrossRef]
- Berglund, B. Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. Infection ecology & epidemiology 2015, 5, 28564. [Google Scholar]
- Gasparrini, A.J.; Wang, B.; Sun, X.; Kennedy, E.A.; Hernandez-Leyva, A.; Ndao, I.M.; Tarr, P.I.; Warner, B.B.; Dantas, G. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nature microbiology 2019, 4, 2285–2297. [Google Scholar] [CrossRef]
- Sheedy, D.B.; Okello, E.; Williams, D.R.; Precht, K.; Cella, E.; Lehenbauer, T.W.; Aly, S.S. Effect of antimicrobial treatment on the dynamics of ceftiofur resistance in Enterobacteriaceae from adult California dairy cows. Microorganisms 2021, 9, 828. [Google Scholar] [CrossRef]
- Jiang, X.; Yang, H.; Dettman, B.; Doyle, M.P. Analysis of fecal microbial flora for antibiotic resistance in ceftiofur-treated calves. Foodbourne Pathogens & Disease 2006, 3, 355–365. [Google Scholar]
- Weinroth, M.D.; Scott, H.M.; Norby, B.; Loneragan, G.H.; Noyes, N.R.; Rovira, P.; Doster, E.; Yang, X.; Woerner, D.R.; Morley, P.S. Effects of ceftiofur and chlortetracycline on the resistomes of feedlot cattle. Applied and environmental microbiology 2018, 84, e00610–00618. [Google Scholar] [CrossRef]
- Dong, L.; Meng, L.; Liu, H.; Wu, H.; Schroyen, M.; Zheng, N.; Wang, J. Effect of Cephalosporin Treatment on the Microbiota and Antibiotic Resistance Genes in Feces of Dairy Cows with Clinical Mastitis. Antibiotics 2022, 11, 117. [Google Scholar] [CrossRef] [PubMed]
- Singer, R.S.; Patterson, S.K.; Wallace, R.L. Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Applied and Environmental Microbiology 2008, 74, 6956–6962. [Google Scholar] [CrossRef]
- Gibson, M.K.; Crofts, T.S.; Dantas, G. Antibiotics and the developing infant gut microbiota and resistome. Current opinion in microbiology 2015, 27, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. Babraham bioinformatics-FastQC a quality control tool for high throughput sequence data. 2010. Available online: https://www. bioinformatics. babraham. ac. uk/projects/fastqc.
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC bioinformatics 2008, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lindgreen, S.; Adair, K.L.; Gardner, P.P. An evaluation of the accuracy and speed of metagenome analysis tools. Scientific reports 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Keegan, K.P.; Glass, E.M.; Meyer, F. MG-RAST, a metagenomics service for analysis of microbial community structure and function. In Microbial environmental genomics (MEG); Springer, 2016; pp. 207–233. [Google Scholar]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic acids research 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome biology 2011, 12, 1–18. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




