Submitted:
24 April 2025
Posted:
25 April 2025
Read the latest preprint version here
Abstract
Keywords:
Introduction
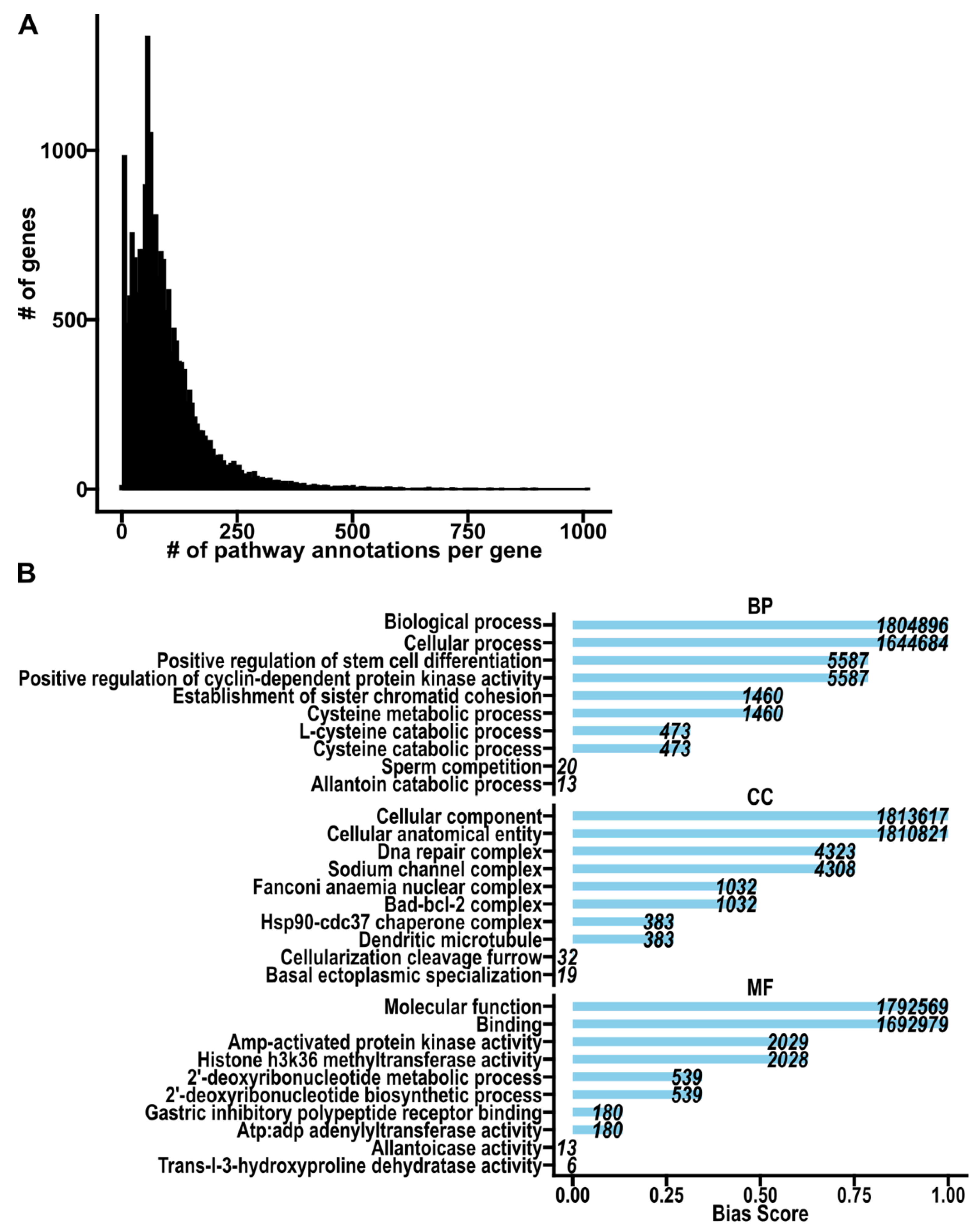
- Key Challenges to Pathway Analysis
- Pathway Annotation
- Visualizing Pathway Findings
- Limitations to Pathway Analysis Utility
- Discrepancies in Molecular Biology Mislead Validation
- Methods for Pathway Analysis Interpretation
- Semantic Similarity Based Methods
- Network Based Methods
- Embedding Based Methods
- Applications of Tools for Pathway Interpretation
- Choosing the Right Tool for Your Research
- Conclusions & Future Directions
| Tool | Year | Method | Access | Database | Visualization | Description |
| REVIGO [91] | 2011 | Semantic | Web | GO | Scatterplots, interactive graph, tree maps | Summarizes GO term lists using semantic similarity and clustering |
| clusterProfiler [94] | 2013 | Semantic | R package | GO, KEGG, DO | Dot plot | Enrichment analysis for GO/KEGG terms and visualization |
| ReCiPa [58] | 2018 | Semantic | R package | KEGG, Reactome | Data tables | Controls redundancy in pathway databases |
| GOGO [93] | 2018 | Semantic | Web, Perl | GO | Data tables | Calculates semantic similarity of GO terms using improved algorithms |
| FunSet [129] | 2019 | Semantic | Web, Standalone | GO | 2D plots | Performs GO enrichment analysis with interactive visualizations |
| GeneSetCluster [130] | 2020 | Semantic | R package | Any | Network graph, dendogram, heatmap | Groups gene-sets post-analysis based on shared genes |
| GOMCL [131] | 2020 | Semantic | Python | GO | Heatmap, Network graph | Clusters GO terms using Markov clustering algorithm |
| GoSemSim [132] | 2020 | Semantic | R package | GO | Data tables | Computes semantic similarity among GO terms for comparison |
| GO-FIGURE! [15] | 2021 | Semantic | Python | GO | Scatterplot | Visualizes GO term similarity with custom scatterplots |
| SimplifyEnrichment [133] | 2022 | Semantic | R package | GO | Heatmap | Clusters with a unique binary cut algorithm. |
| RICHNET [98] | 2019 | Network | R protocol | MSigDB | Network graph | Automated gene-set network creation |
| EnrichmentMap [66] | 2019 | Network | Cytoscape | Any | Interactive network | Detailed enrichment mapping |
| Gscluster [99] | 2019 | Network | Web, R Package | MSigDB | Interactive network | Network-weighted gene-set clustering integrating PPI data |
| aPEAR [101] | 2019 | Network | R package | Any | Network graph | Clustering with automated naming |
| GeneFEAST [100] | 2023 | Network | Web, Python | Any | Heatmap, Dot plot, Upset plot | Highlights multi-enrichment genes |
| vissE [102,103] | 2023 | Network | R package | MSigDB, Any | Network graph | Visualizes higher-order interactions |
| pathlinkR [97] | 2024 | Network | R package | Reactome, MSigDB, InnateDB | Network graph, Volcano plot, Dot plot | Integrated PPI network construction |
| PAVER [110] | 2024 | Embedding | Web, R package | Any | UMAP, Heatmap, Dot plot | Embedding-based clustering with UMAP for clear pathway visualization. |
| Mondrian-Map [113] | 2024 | Embedding | Python | WikiPathways | Mondrian Map | Embedding visualizations highlighting pathway interactions and crosstalk. |
| GOsummaries [134] | 2015 | Word Cloud | R package | GO | PCA, Boxplot | Visualizes GO analyses as word clouds and overlays results. |
| genesetSV [135] | 2023 | Game Theory | Python | KEGG, MSigDB | Scatterplot | Uses Shapley values for ranking and reducing pathway sets. |
Acknowledgements
Glossary
References
- Manzoni, C.; et al. Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences. Briefings in Bioinformatics 2016, 19, 286–302. [Google Scholar] [CrossRef] [PubMed]
- Veenstra, T.D. , Omics in Systems Biology: Current Progress and Future Outlook. Proteomics 2021, 21, e2000235. [Google Scholar] [CrossRef] [PubMed]
- Herr, T.M.; et al. A conceptual model for translating omic data into clinical action. Journal of Pathology Informatics 2015, 6, 46. [Google Scholar] [CrossRef]
- García-Campos, M.A., J. Espinal-Enríquez, and E. Hernández-Lemus, Pathway Analysis: State of the Art. Front Physiol 2015, 6, 383.
- Wegman-Points, L.; et al. Subcellular partitioning of protein kinase activity revealed by functional kinome profiling. Scientific Reports 2022, 12, 17300. [Google Scholar] [CrossRef]
- Ramanan, V.K.; et al. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet 2012, 28, 323–332. [Google Scholar] [CrossRef]
- Krassowski, M.; et al. State of the Field in Multi-Omics Research: From Computational Needs to Data Mining and Sharing. Front Genet 2020, 11, 610798. [Google Scholar] [CrossRef]
- Sboner, A.; et al. The real cost of sequencing: higher than you think! Genome Biol 2011, 12, 125. [Google Scholar] [CrossRef]
- D'Adamo, G.L., J.T. Widdop, and E.M. Giles, The future is now? Clinical and translational aspects of "Omics" technologies. Immunol Cell Biol 2021, 99, 168–176. [CrossRef]
- Denecker, T. and G. Lelandais, Omics Analyses: How to Navigate Through a Constant DataData Deluge, in Yeast Functional Genomics: Methods and Protocols, F. Devaux, Editor. 2022, Springer US: New York, NY. p. 457-471.
- Bell, G., T. Hey, and A. Szalay, Computer science. Beyond the data deluge. Science 2009, 323, 1297–1298.
- Stead, W.W.; et al. Biomedical informatics: changing what physicians need to know and how they learn. Acad Med 2011, 86, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Pita-Juárez, Y.; et al. The Pathway Coexpression Network: Revealing pathway relationships. PLOS Computational Biology 2018, 14, e1006042. [Google Scholar] [CrossRef] [PubMed]
- Chicco, D. and G. Agapito, Nine quick tips for pathway enrichment analysis. 2022, 18, e1010348.
- Reijnders, M.J. and R.M. Waterhouse, Summary visualizations of gene ontology terms with GO-Figure! Frontiers in Bioinformatics 2021, 1, 6.
- Yu, C.; et al. A strategy for evaluating pathway analysis methods. BMC Bioinformatics 2017, 18, 453. [Google Scholar] [CrossRef]
- Searson, P.C. , The Cancer Moonshot, the role of in vitro models, model accuracy, and the need for validation. Nature Nanotechnology 2023, 18, 1121–1123. [Google Scholar] [CrossRef]
- Durinikova, E., K. Buzo, and S. Arena, Preclinical models as patients’ avatars for precision medicine in colorectal cancer: past and future challenges. Journal of Experimental & Clinical Cancer Research 2021, 40, 185.
- Diaz-Uriarte, R.; et al. Ten quick tips for biomarker discovery and validation analyses using machine learning. PLOS Computational Biology 2022, 18, e1010357. [Google Scholar] [CrossRef]
- Grabowski, T. , et al. Between Biological Relevancy and Statistical Significance - Step for Assessment Harmonization. 2021.
- Committee, E.S.; et al. Guidance on the assessment of the biological relevance of data in scientific assessments. EFSA Journal 2017, 15, e04970. [Google Scholar]
- Perez-Riverol, Y.; et al. Quantifying the impact of public omics data. Nat Commun 2019, 10, 3512. [Google Scholar] [CrossRef]
- Misra, B.B.; et al. Integrated omics: tools, advances and future approaches. Journal of Molecular Endocrinology 2019, 62, R21–R45. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Fernández, D.; et al. PathMe: merging and exploring mechanistic pathway knowledge. BMC Bioinformatics 2019, 20, 243. [Google Scholar] [CrossRef] [PubMed]
- Wieder, C.; et al. PathIntegrate: Multivariate modelling approaches for pathway-based multi-omics data integration. PLOS Computational Biology 2024, 20, e1011814. [Google Scholar] [CrossRef]
- Canzler, S. and J. Hackermüller, multiGSEA: a GSEA-based pathway enrichment analysis for multi-omics data. BMC Bioinformatics 2020, 21, 561.
- Ivanisevic, T. and R.N. Sewduth, Multi-Omics Integration for the Design of Novel Therapies and the Identification of Novel Biomarkers. Proteomes 2023, 11, 34. [CrossRef]
- Mohr, A.E.; et al. Navigating Challenges and Opportunities in Multi-Omics Integration for Personalized Healthcare. Biomedicines 2024, 12, 1496. [Google Scholar] [CrossRef]
- Conroy, G. Retractions caused by honest mistakes are extremely stressful, say researchers. Nature 2025. [Google Scholar] [CrossRef]
- Kovacs, M.; et al. Opening the black box of article retractions: exploring the causes and consequences of data management errors. R Soc Open Sci 2024, 11, 240844. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016, 3, 160018. [Google Scholar] [CrossRef]
- Doniparthi, G., T. Mühlhaus, and S. Deßloch, Integrating FAIR Experimental Metadata for Multi-omics Data Analysis. Datenbank-Spektrum 2024, 24, 107–115. [CrossRef]
- Jan, M.; et al. A multi-omics digital research object for the genetics of sleep regulation. Scientific Data 2019, 6, 258. [Google Scholar] [CrossRef] [PubMed]
- Khatri, P., M. Sirota, and A.J. Butte, Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol 2012, 8, e1002375.
- Nguyen, T.-M.; et al. Identifying significantly impacted pathways: a comprehensive review and assessment. Genome Biology 2019, 20, 203. [Google Scholar] [CrossRef]
- Nam, D. and S.Y. Kim, Gene-set approach for expression pattern analysis. Brief Bioinform 2008, 9, 189–197. [CrossRef]
- Maghsoudi, Z.; et al. A comprehensive survey of the approaches for pathway analysis using multi-omics data integration. Briefings in Bioinformatics 2022, 23. [Google Scholar] [CrossRef]
- García-Campos, M.A., J. Espinal-Enríquez, and E. Hernández-Lemus, Pathway Analysis: State of the Art. Frontiers in Physiology 2015, 6.
- Winston, J.E., Twenty-First Century Biological Nomenclature—The Enduring Power of Names. Integrative and Comparative Biology 2018, 58, 1122–1131.
- Vassalli, P. , The pathophysiology of tumor necrosis factors. Annu Rev Immunol 1992, 10, 411–452. [Google Scholar] [CrossRef]
- Webster, J.D. and D. Vucic, The Balance of TNF Mediated Pathways Regulates Inflammatory Cell Death Signaling in Healthy and Diseased Tissues. Front Cell Dev Biol 2020, 8, 365.
- Wang, D.; et al. Designing Theory-Driven User-Centric Explainable AI, in Proceedings of the 2019 CHI Conference on Human Factors in Computing Systems. 2019, Association for Computing Machinery: Glasgow, Scotland Uk. p. Paper 601.
- Jara, J.H.; et al. Tumor necrosis factor alpha stimulates NMDA receptor activity in mouse cortical neurons resulting in ERK-dependent death. J Neurochem 2007, 100, 1407–1420. [Google Scholar] [CrossRef]
- Sebastian-Leon, P.; et al. Understanding disease mechanisms with models of signaling pathway activities. BMC Systems Biology 2014, 8, 121. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; et al. Prioritizing biological pathways by recognizing context in time-series gene expression data. BMC Bioinformatics 2016, 17, 477. [Google Scholar] [CrossRef] [PubMed]
- Sjöström, J. and J. Bergh, How apoptosis is regulated, and what goes wrong in cancer. BMJ 2001, 322, 1538–1539. [CrossRef]
- Nguyen, T.T.M., G. Gillet, and N. Popgeorgiev, Caspases in the Developing Central Nervous System: Apoptosis and Beyond. Frontiers in Cell and Developmental Biology 2021, 9.
- Ryu, J.R.; et al. Control of adult neurogenesis by programmed cell death in the mammalian brain. Molecular Brain 2016, 9, 43. [Google Scholar] [CrossRef]
- Anosike, N.L.; et al. Necroptosis in the developing brain: role in neurodevelopmental disorders. Metabolic Brain Disease 2023, 38, 831–837. [Google Scholar] [CrossRef]
- Saini, S., P. Kakati, and K. Singh, Role of Inflammation in Tissue Regeneration and Repair, in Inflammation Resolution and Chronic Diseases, A. Tripathi, et al., Editors. 2024, Springer Nature Singapore: Singapore. p. 103-127.
- Choi, B., C. Lee, and J.-W. Yu, Distinctive role of inflammation in tissue repair and regeneration. Archives of Pharmacal Research 2023, 46, 78–89. [CrossRef]
- Wyss-Coray, T. and L. Mucke, Inflammation in neurodegenerative disease--a double-edged sword. Neuron 2002, 35, 419–432. [CrossRef]
- Gasque, P.; et al. Roles of the complement system in human neurodegenerative disorders: pro-inflammatory and tissue remodeling activities. Mol Neurobiol 2002, 25, 1–17. [Google Scholar] [CrossRef]
- Shih, R.-H., C.-Y. Wang, and C.-M. Yang, NF-kappaB Signaling Pathways in Neurological Inflammation: A Mini Review. Frontiers in Molecular Neuroscience 2015, 8.
- Sun, S.-C. , The non-canonical NF-κB pathway in immunity and inflammation. Nature Reviews Immunology 2017, 17, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Adriaens, M.E.; et al. The public road to high-quality curated biological pathways. Drug Discovery Today 2008, 13, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.-G. and A.R. Pico, Using published pathway figures in enrichment analysis and machine learning. BMC Genomics 2023, 24, 713.
- Vivar, J.C.; et al. Redundancy control in pathway databases (ReCiPa): an application for improving gene-set enrichment analysis in Omics studies and "Big data" biology. Omics 2013, 17, 414–422. [Google Scholar] [CrossRef]
- Pastrello, C., Y. Niu, and I. Jurisica, Pathway Enrichment Analysis of Microarray Data. Methods Mol Biol 2022, 2401, 147–159.
- Gable, A.L.; et al. Systematic assessment of pathway databases, based on a diverse collection of user-submitted experiments. Briefings in Bioinformatics 2022, 23. [Google Scholar] [CrossRef]
- Maertens, A.; et al. Functionally Enigmatic Genes in Cancer: Using TCGA Data to Map the Limitations of Annotations. Sci Rep 2020, 10, 4106. [Google Scholar] [CrossRef]
- Stoney, R.A.; et al. Using set theory to reduce redundancy in pathway sets. BMC Bioinformatics 2018, 19, 386. [Google Scholar] [CrossRef]
- Krassowski, M.; et al. State of the Field in Multi-Omics Research: From Computational Needs to Data Mining and Sharing. Front Genet 2020, 11, 610798. [Google Scholar] [CrossRef]
- Hanspers, K.; et al. Ten simple rules for creating reusable pathway models for computational analysis and visualization. PLOS Computational Biology 2021, 17, e1009226. [Google Scholar] [CrossRef]
- He, C.; et al. Interactive visual facets to support fluid exploratory search. Journal of Visualization 2023, 26, 211–230. [Google Scholar] [CrossRef]
- Reimand, J.; et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nature Protocols 2019, 14, 482–517. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, S. and S. Anders, Exploring dimension-reduced embeddings with Sleepwalk. Genome Res 2020, 30, 749–756. [CrossRef]
- Li, Y.; et al. Exaggerated false positives by popular differential expression methods when analyzing human population samples. Genome Biology 2022, 23, 79. [Google Scholar] [CrossRef]
- Radulescu, E.; et al. Identification and prioritization of gene sets associated with schizophrenia risk by co-expression network analysis in human brain. Mol Psychiatry 2020, 25, 791–804. [Google Scholar] [CrossRef]
- Sapienza, J.; et al. Importance of the dysregulation of the kynurenine pathway on cognition in schizophrenia: a systematic review of clinical studies. Eur Arch Psychiatry Clin Neurosci 2023, 273, 1317–1328. [Google Scholar] [CrossRef]
- Rusina, P.V.; et al. Genetic support for FDA-approved drugs over the past decade. Nat Rev Drug Discov 2023, 22, 864. [Google Scholar] [CrossRef]
- Ochoa, D.; et al. Human genetics evidence supports two-thirds of the 2021 FDA-approved drugs. Nat Rev Drug Discov 2022, 21, 551. [Google Scholar] [CrossRef]
- Diogo, D.; et al. TYK2 protein-coding variants protect against rheumatoid arthritis and autoimmunity, with no evidence of major pleiotropic effects on non-autoimmune complex traits. PLoS One 2015, 10, e0122271. [Google Scholar] [CrossRef]
- MacNamara, A.; et al. Network and pathway expansion of genetic disease associations identifies successful drug targets. Sci Rep 2020, 10, 20970. [Google Scholar] [CrossRef]
- de la Fuente van Bentem, S.; et al. Towards functional phosphoproteomics by mapping differential phosphorylation events in signaling networks. PROTEOMICS 2008, 8, 4453–4465. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, E.A.; et al. Workability of mRNA Sequencing for Predicting Protein Abundance. Genes 2023, 14, 2065. [Google Scholar] [CrossRef] [PubMed]
- Prabahar, A.; et al. Unraveling the complex relationship between mRNA and protein abundances: a machine learning-based approach for imputing protein levels from RNA-seq data. NAR Genomics and Bioinformatics 2024, 6. [Google Scholar] [CrossRef] [PubMed]
- de Sousa Abreu, R.; et al. Global signatures of protein and mRNA expression levels. Molecular BioSystems 2009, 5, 1512–1526. [Google Scholar] [CrossRef]
- Upadhya, S.R. and C.J. Ryan, Experimental reproducibility limits the correlation between mRNA and protein abundances in tumor proteomic profiles. Cell Rep Methods 2022, 2, 100288. [CrossRef]
- Arshad, O.A.; et al. An Integrative Analysis of Tumor Proteomic and Phosphoproteomic Profiles to Examine the Relationships Between Kinase Activity and Phosphorylation*. Molecular & Cellular Proteomics 2019, 18 (Supplement 1), S26-S36.
- Liu, Y., A. Beyer, and R. Aebersold, On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [CrossRef]
- Handly, L.N., J. Yao, and R. Wollman, Signal Transduction at the Single-Cell Level: Approaches to Study the Dynamic Nature of Signaling Networks. J Mol Biol 2016, 428, 3669–3682. [CrossRef]
- Creeden, J.F.; et al. Kinome Array Profiling of Patient-Derived Pancreatic Ductal Adenocarcinoma Identifies Differentially Active Protein Tyrosine Kinases. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef]
- Litichevskiy, L.; et al. A Library of Phosphoproteomic and Chromatin Signatures for Characterizing Cellular Responses to Drug Perturbations. Cell Syst 2018, 6, 424–443.e7. [Google Scholar] [CrossRef]
- Reinecke, M.; et al. Kinobeads: A Chemical Proteomic Approach for Kinase Inhibitor Selectivity Profiling and Target Discovery, in Target Discovery and Validation. 2019. p. 97-130.
- Patricelli, M.P.; et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem Biol 2011, 18, 699–710. [Google Scholar] [CrossRef]
- Alganem, K.; et al. The active kinome: The modern view of how active protein kinase networks fit in biological research. Curr Opin Pharmacol 2022, 62, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.; et al. Network propagation: a universal amplifier of genetic associations. Nat Rev Genet 2017, 18, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Supek, F. and N. Skunca, Visualizing GO Annotations. Methods Mol Biol 2017, 1446, 207–220.
- Gan, M., X. Dou, and R. Jiang, From ontology to semantic similarity: calculation of ontology-based semantic similarity. ScientificWorldJournal 2013, 2013, 793091. [CrossRef]
- Supek, F.; et al. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 2011, 6, e21800. [Google Scholar] [CrossRef]
- Pesquita, C.; et al. Semantic similarity in biomedical ontologies. PLoS Comput Biol 2009, 5, e1000443. [Google Scholar] [CrossRef]
- Zhao, C. and Z. Wang, GOGO: An improved algorithm to measure the semantic similarity between gene ontology terms. Sci Rep 2018, 8, 15107. [CrossRef]
- Yu, G.; et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Galeota, E., K. Kishore, and M. Pelizzola, Ontology-driven integrative analysis of omics data through Onassis. Sci Rep 2020, 10, 703. [CrossRef]
- Duong, D.; et al. Word and Sentence Embedding Tools to Measure Semantic Similarity of Gene Ontology Terms by Their Definitions. J Comput Biol 2019, 26, 38–52. [Google Scholar] [CrossRef]
- Blimkie, T.M., A. An, and R.E.W. Hancock, Facilitating pathway and network based analysis of RNA-Seq data with pathlinkR. PLOS Computational Biology 2024, 20, e1012422.
- Prummer, M. , Enhancing gene set enrichment using networks. F1000Res 2019, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; et al. GScluster: network-weighted gene-set clustering analysis. BMC Genomics 2019, 20, 352. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; et al. GeneFEAST: the pivotal, gene-centric step in functional enrichment analysis interpretation. arXiv 2023, arXiv:2309.00061. [Google Scholar] [CrossRef]
- Kerseviciute, I. and J. Gordevicius, aPEAR: an R package for autonomous visualization of pathway enrichment networks. Bioinformatics 2023, 39. [Google Scholar] [CrossRef]
- Bhuva, D.D.; et al. vissE: a versatile tool to identify and visualise higher-order molecular phenotypes from functional enrichment analysis. BMC Bioinformatics 2024, 25, 64. [Google Scholar] [CrossRef]
- Mohamed, A.; et al. vissE.cloud: a webserver to visualise higher order molecular phenotypes from enrichment analysis. Nucleic Acids Res 2023, 51, W593–W600. [Google Scholar] [CrossRef]
- Major, V., A. Surkis, and Y. Aphinyanaphongs, Utility of General and Specific Word Embeddings for Classifying Translational Stages of Research. AMIA Annu Symp Proc 2018, 2018, 1405–1414.
- Chiu, B., et al. How to train good word embeddings for biomedical NLP. in Proceedings of the 15th workshop on biomedical natural language processing. 2016.
- Mikolov, T.; et al. Efficient estimation of word representations in vector space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Ofer, D., N. Brandes, and M. Linial, The language of proteins: NLP, machine learning & protein sequences. Comput Struct Biotechnol J 2021, 19, 1750–1758.
- Xenos, A.; et al. Linear functional organization of the omic embedding space. Bioinformatics 2021, 37, 3839–3847. [Google Scholar] [CrossRef] [PubMed]
- Asgari, E. and M.R. Mofrad, Continuous Distributed Representation of Biological Sequences for Deep Proteomics and Genomics. PLoS One 2015, 10, e0141287.
- Ryan, V.W.; et al. Interpreting and visualizing pathway analyses using embedding representations with PAVER. Bioinformation 2024, 20, 700–704. [Google Scholar] [CrossRef]
- Kulmanov, M.; et al. Semantic similarity and machine learning with ontologies. Brief Bioinform 2021, 22. [Google Scholar] [CrossRef]
- Lerman, G. and B.E. Shakhnovich, Defining functional distance using manifold embeddings of gene ontology annotations. Proc Natl Acad Sci U S A 2007, 104, 11334–11339. [CrossRef]
- Al Abir, F. and J.Y. Chen, Mondrian Abstraction and Language Model Embeddings for Differential Pathway Analysis. bioRxiv 2024. [Google Scholar]
- Chen, D.-Q.; et al. Identification of Differentially Expressed Genes and Signaling Pathways in Acute Myocardial Infarction Based on Integrated Bioinformatics Analysis. Cardiovascular Therapeutics 2019, 2019, 8490707. [Google Scholar] [CrossRef]
- Jia, R.; et al. Identification of key genes unique to the luminal a and basal-like breast cancer subtypes via bioinformatic analysis. World Journal of Surgical Oncology 2020, 18, 268. [Google Scholar] [CrossRef]
- Niu, Y.; et al. Bioinformatics to analyze the differentially expressed genes in different degrees of Alzheimer’s disease and their roles in progress of the disease. Journal of Applied Genetics 2024. [Google Scholar] [CrossRef]
- Gamazon, E.R.; et al. Multi-tissue transcriptome analyses identify genetic mechanisms underlying neuropsychiatric traits. Nature Genetics 2019, 51, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Kulasinghe, A.; et al. Transcriptomic profiling of cardiac tissues from SARS-CoV-2 patients identifies DNA damage. Immunology 2023, 168, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Dalit, L.; et al. Divergent cytokine and transcriptional signatures control functional T follicular helper cell heterogeneity. bioRxiv 2024, 2024.06.12.598622.
- Lee, J.-Y.; et al. Inhibition of HTR2B-mediated serotonin signaling in colorectal cancer suppresses tumor growth through ERK signaling. Biomedicine & Pharmacotherapy 2024, 179, 117428. [Google Scholar]
- Nguyen, J.H.; et al. Developmental pyrethroid exposure disrupts molecular pathways for MAP kinase and circadian rhythms in mouse brain. bioRxiv 2024. [Google Scholar]
- Curtis, M.A.; et al. Developmental pyrethroid exposure in mouse leads to disrupted brain metabolism in adulthood. NeuroToxicology 2024, 103, 87–95. [Google Scholar] [CrossRef]
- O'Donovan, S.; et al. Shared and unique transcriptional changes in the orbitofrontal cortex in psychiatric disorders and suicide. Translation: The University of Toledo Journal of Medical Sciences 2024, 12. [Google Scholar] [CrossRef]
- Hu, Y.; et al. Probiotic Protects Kidneys Exposed to Microcystin-LR. Translation: The University of Toledo Journal of Medical Sciences 2024, 12. [Google Scholar] [CrossRef]
- Hodgman, C., A. French, and D. Westhead, BIOS Instant Notes in Bioinformatics. 2009: Taylor & Francis.
- Karp, P.D.; et al. Pathway size matters: the influence of pathway granularity on over-representation (enrichment analysis) statistics. BMC Genomics 2021, 22, 191. [Google Scholar] [CrossRef]
- Wijesooriya, K.; et al. Urgent need for consistent standards in functional enrichment analysis. PLoS Comput Biol 2022, 18, e1009935. [Google Scholar] [CrossRef]
- Ziemann, M., B. Schroeter, and A. Bora, Two subtle problems with over-representation analysis. Bioinformatics Advances 2024.
- Hale, M.L., I. Thapa, and D. Ghersi, FunSet: an open-source software and web server for performing and displaying Gene Ontology enrichment analysis. BMC Bioinformatics 2019, 20, 359. [Google Scholar] [CrossRef] [PubMed]
- Ewing, E.; et al. GeneSetCluster: a tool for summarizing and integrating gene-set analysis results. BMC Bioinformatics 2020, 21, 443. [Google Scholar] [CrossRef] [PubMed]
- Wang, G., D.H. Oh, and M. Dassanayake, GOMCL: a toolkit to cluster, evaluate, and extract non-redundant associations of Gene Ontology-based functions. BMC Bioinformatics 2020, 21, 139.
- Yu, G. , Gene Ontology Semantic Similarity Analysis Using GOSemSim. Methods Mol Biol 2020, 2117, 207–215. [Google Scholar]
- Gu, Z. and D. Hübschmann, SimplifyEnrichment: A Bioconductor Package for Clustering and Visualizing Functional Enrichment Results. Genomics, Proteomics & Bioinformatics 2022, 21, 190–202. [Google Scholar]
- Kolde, R. and J. Vilo, GOsummaries: an R Package for Visual Functional Annotation of Experimental Data. F1000Res 2015, 4, 574. [Google Scholar] [CrossRef]
- Balestra, C.; et al. Redundancy-aware unsupervised ranking based on game theory: Ranking pathways in collections of gene sets. PLoS One 2023, 18, e0282699. [Google Scholar] [CrossRef]
| Gene | # of Pathways |
|---|---|
| TGFB1 transforming growth factor beta 1 | 1010 |
| CTNNB1 catenin beta 1 | 894 |
| ACADL acyl-CoA dehydrogenase long chain | 120 |
| ACTBL2 actin beta like 2 | 120 |
| ABCA6 ATP binding cassette subfamily A member 6 | 72 |
| ACKR1 atypical chemokine receptor 1 (Duffy blood group) | 72 |
| ABCF3 ATP binding cassette subfamily F member 3 | 44 |
| ADISSP adipose secreted signaling protein | 44 |
| C6orf62 chromosome 6 open reading frame 62 | 2 |
| CTAGE3P CTAGE family member 3, pseudogene | 2 |
| Locus Type | Count |
| pseudogene | 13940 |
| RNA, long non-coding | 5640 |
| RNA, micro | 1912 |
| gene with protein product | 611 |
| RNA, transfer | 591 |
| RNA, small nucleolar | 568 |
| immunoglobulin pseudogene | 202 |
| readthrough | 143 |
| RNA, cluster | 119 |
| fragile site | 116 |
| endogenous retrovirus | 92 |
| T cell receptor gene | 67 |
| RNA, ribosomal | 58 |
| immunoglobulin gene | 55 |
| RNA, small nuclear | 51 |
| region | 46 |
| unknown | 46 |
| T cell receptor pseudogene | 38 |
| RNA, misc | 29 |
| virus integration site | 8 |
| complex locus constituent | 6 |
| RNA, vault | 4 |
| RNA, Y | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





